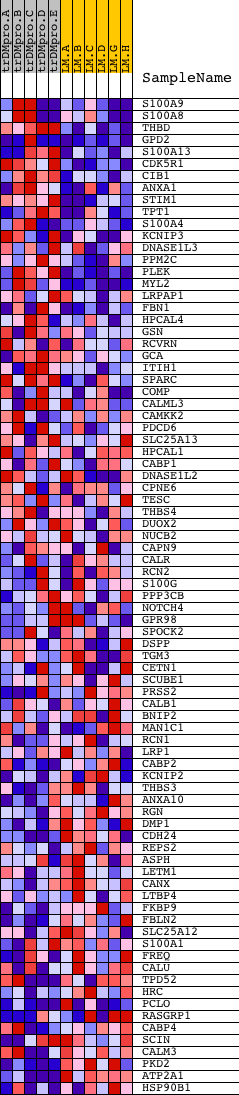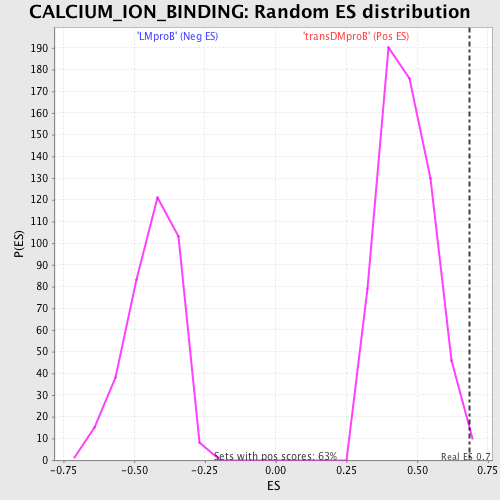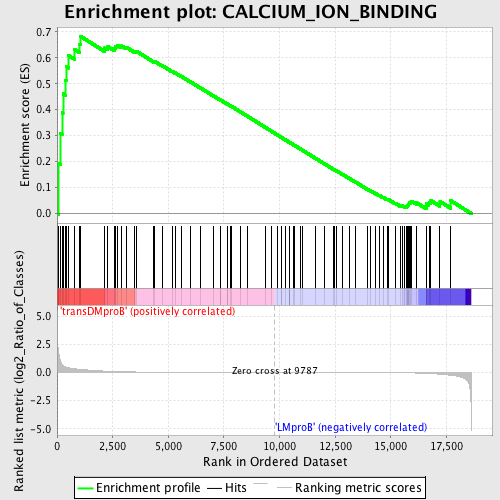
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | CALCIUM_ION_BINDING |
| Enrichment Score (ES) | 0.6833185 |
| Normalized Enrichment Score (NES) | 1.4812046 |
| Nominal p-value | 0.0031695722 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | S100A9 | 9774 | 64 | 1.652 | 0.1925 | Yes | ||
| 2 | S100A8 | 15527 | 156 | 1.021 | 0.3087 | Yes | ||
| 3 | THBD | 14404 | 224 | 0.708 | 0.3891 | Yes | ||
| 4 | GPD2 | 4773 9014 | 266 | 0.627 | 0.4613 | Yes | ||
| 5 | S100A13 | 15532 | 389 | 0.485 | 0.5122 | Yes | ||
| 6 | CDK5R1 | 20745 | 411 | 0.466 | 0.5664 | Yes | ||
| 7 | CIB1 | 17780 | 523 | 0.402 | 0.6081 | Yes | ||
| 8 | ANXA1 | 9282 | 773 | 0.320 | 0.6326 | Yes | ||
| 9 | STIM1 | 18164 | 1005 | 0.276 | 0.6530 | Yes | ||
| 10 | TPT1 | 5799 | 1038 | 0.270 | 0.6833 | Yes | ||
| 11 | S100A4 | 9773 | 2144 | 0.118 | 0.6377 | No | ||
| 12 | KCNIP3 | 14441 | 2282 | 0.105 | 0.6427 | No | ||
| 13 | DNASE1L3 | 21925 | 2570 | 0.084 | 0.6372 | No | ||
| 14 | PPM2C | 491 | 2625 | 0.080 | 0.6438 | No | ||
| 15 | PLEK | 7072 | 2693 | 0.075 | 0.6491 | No | ||
| 16 | MYL2 | 16713 | 2900 | 0.063 | 0.6455 | No | ||
| 17 | LRPAP1 | 5005 | 3114 | 0.053 | 0.6402 | No | ||
| 18 | FBN1 | 14449 | 3473 | 0.040 | 0.6257 | No | ||
| 19 | HPCAL4 | 16096 | 3570 | 0.037 | 0.6249 | No | ||
| 20 | GSN | 2784 | 4352 | 0.021 | 0.5853 | No | ||
| 21 | RCVRN | 20835 | 4395 | 0.021 | 0.5855 | No | ||
| 22 | GCA | 2813 15003 10720 7830 | 4740 | 0.017 | 0.5691 | No | ||
| 23 | ITIH1 | 21895 | 5167 | 0.014 | 0.5477 | No | ||
| 24 | SPARC | 20444 | 5304 | 0.013 | 0.5419 | No | ||
| 25 | COMP | 18589 | 5585 | 0.012 | 0.5282 | No | ||
| 26 | CALML3 | 21553 | 5974 | 0.010 | 0.5084 | No | ||
| 27 | CAMKK2 | 9870 | 6438 | 0.008 | 0.4843 | No | ||
| 28 | PDCD6 | 21404 | 7047 | 0.006 | 0.4522 | No | ||
| 29 | SLC25A13 | 17221 1159 | 7331 | 0.005 | 0.4376 | No | ||
| 30 | HPCAL1 | 21307 2123 | 7364 | 0.005 | 0.4364 | No | ||
| 31 | CABP1 | 16412 | 7636 | 0.004 | 0.4223 | No | ||
| 32 | DNASE1L2 | 960 23092 | 7670 | 0.004 | 0.4211 | No | ||
| 33 | CPNE6 | 22010 | 7804 | 0.004 | 0.4143 | No | ||
| 34 | TESC | 16725 | 7843 | 0.004 | 0.4128 | No | ||
| 35 | THBS4 | 21393 | 8254 | 0.003 | 0.3910 | No | ||
| 36 | DUOX2 | 14453 | 8548 | 0.002 | 0.3755 | No | ||
| 37 | NUCB2 | 18114 | 9371 | 0.001 | 0.3312 | No | ||
| 38 | CAPN9 | 3772 18422 | 9638 | 0.000 | 0.3169 | No | ||
| 39 | CALR | 18814 | 9917 | -0.000 | 0.3019 | No | ||
| 40 | RCN2 | 19438 | 10067 | -0.001 | 0.2939 | No | ||
| 41 | S100G | 24016 | 10077 | -0.001 | 0.2935 | No | ||
| 42 | PPP3CB | 5285 | 10274 | -0.001 | 0.2831 | No | ||
| 43 | NOTCH4 | 23277 9475 | 10427 | -0.001 | 0.2750 | No | ||
| 44 | GPR98 | 21400 | 10440 | -0.001 | 0.2745 | No | ||
| 45 | SPOCK2 | 13585 | 10447 | -0.001 | 0.2743 | No | ||
| 46 | DSPP | 8867 | 10603 | -0.002 | 0.2662 | No | ||
| 47 | TGM3 | 10168 | 10668 | -0.002 | 0.2629 | No | ||
| 48 | CETN1 | 6443 | 10938 | -0.002 | 0.2486 | No | ||
| 49 | SCUBE1 | 7231 | 11024 | -0.002 | 0.2443 | No | ||
| 50 | PRSS2 | 17474 | 11634 | -0.004 | 0.2119 | No | ||
| 51 | CALB1 | 4469 | 12010 | -0.004 | 0.1922 | No | ||
| 52 | BNIP2 | 179 19388 | 12429 | -0.005 | 0.1703 | No | ||
| 53 | MAN1C1 | 10514 6064 15714 | 12478 | -0.006 | 0.1684 | No | ||
| 54 | RCN1 | 9710 | 12578 | -0.006 | 0.1637 | No | ||
| 55 | LRP1 | 9284 | 12835 | -0.007 | 0.1507 | No | ||
| 56 | CABP2 | 23765 | 13140 | -0.008 | 0.1352 | No | ||
| 57 | KCNIP2 | 23655 3725 | 13417 | -0.009 | 0.1213 | No | ||
| 58 | THBS3 | 15542 1796 | 13973 | -0.011 | 0.0927 | No | ||
| 59 | ANXA10 | 18869 | 14085 | -0.012 | 0.0881 | No | ||
| 60 | RGN | 24371 | 14322 | -0.013 | 0.0769 | No | ||
| 61 | DMP1 | 16776 | 14479 | -0.014 | 0.0702 | No | ||
| 62 | CDH24 | 10726 | 14660 | -0.016 | 0.0623 | No | ||
| 63 | REPS2 | 24018 | 14829 | -0.017 | 0.0553 | No | ||
| 64 | ASPH | 2498 2460 2396 2370 2357 2502 2516 2375 2337 12276 2381 2454 7246 2480 2393 | 14886 | -0.018 | 0.0544 | No | ||
| 65 | LETM1 | 7108 16563 12114 | 15202 | -0.022 | 0.0401 | No | ||
| 66 | CANX | 4474 | 15443 | -0.027 | 0.0303 | No | ||
| 67 | LTBP4 | 1739 17918 | 15509 | -0.028 | 0.0302 | No | ||
| 68 | FKBP9 | 17433 | 15620 | -0.032 | 0.0280 | No | ||
| 69 | FBLN2 | 994 8956 | 15713 | -0.035 | 0.0271 | No | ||
| 70 | SLC25A12 | 14562 | 15739 | -0.035 | 0.0300 | No | ||
| 71 | S100A1 | 15271 | 15764 | -0.036 | 0.0330 | No | ||
| 72 | FREQ | 4734 8983 15046 | 15814 | -0.038 | 0.0348 | No | ||
| 73 | CALU | 1124 1149 8683 1162 4472 | 15815 | -0.038 | 0.0394 | No | ||
| 74 | TPD52 | 18830 5794 | 15816 | -0.038 | 0.0439 | No | ||
| 75 | HRC | 18251 | 15893 | -0.041 | 0.0446 | No | ||
| 76 | PCLO | 11207 6495 | 15938 | -0.043 | 0.0474 | No | ||
| 77 | RASGRP1 | 5360 14476 9694 | 16143 | -0.053 | 0.0426 | No | ||
| 78 | CABP4 | 13125 | 16588 | -0.082 | 0.0284 | No | ||
| 79 | SCIN | 21079 | 16596 | -0.083 | 0.0379 | No | ||
| 80 | CALM3 | 8682 | 16715 | -0.094 | 0.0427 | No | ||
| 81 | PKD2 | 16772 | 16797 | -0.102 | 0.0504 | No | ||
| 82 | ATP2A1 | 17639 | 17208 | -0.156 | 0.0468 | No | ||
| 83 | HSP90B1 | 19657 | 17695 | -0.245 | 0.0497 | No |