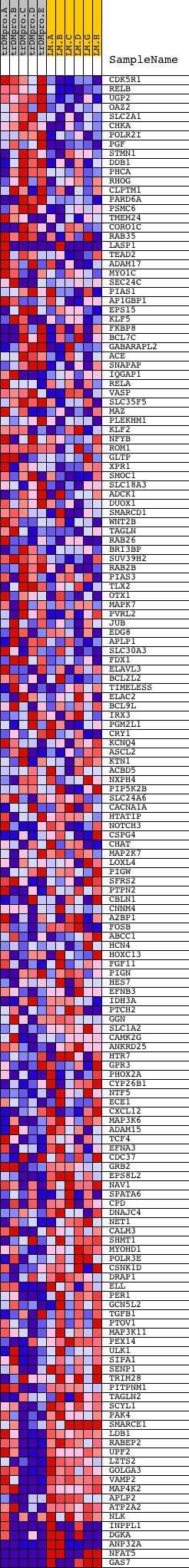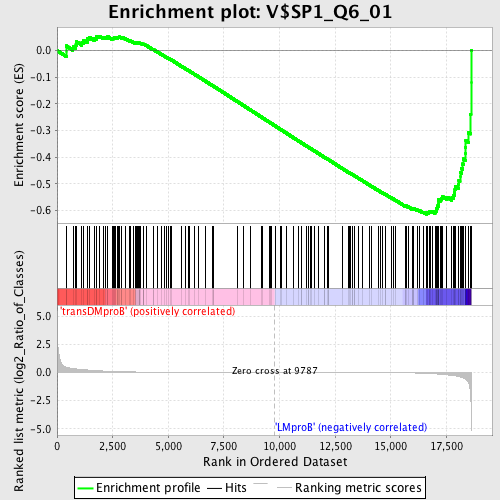
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$SP1_Q6_01 |
| Enrichment Score (ES) | -0.6146496 |
| Normalized Enrichment Score (NES) | -1.5352594 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.5327344 |
| FWER p-Value | 0.804 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDK5R1 | 20745 | 411 | 0.466 | -0.0024 | No | ||
| 2 | RELB | 17942 | 412 | 0.466 | 0.0174 | No | ||
| 3 | UGP2 | 20518 | 714 | 0.339 | 0.0156 | No | ||
| 4 | OAZ2 | 9499 | 848 | 0.305 | 0.0214 | No | ||
| 5 | SLC2A1 | 16107 | 870 | 0.301 | 0.0331 | No | ||
| 6 | CHKA | 23759 | 1115 | 0.256 | 0.0308 | No | ||
| 7 | POLR2I | 12839 | 1179 | 0.244 | 0.0378 | No | ||
| 8 | PGF | 21020 | 1354 | 0.217 | 0.0376 | No | ||
| 9 | STMN1 | 9261 | 1374 | 0.216 | 0.0458 | No | ||
| 10 | DDB1 | 23931 | 1462 | 0.202 | 0.0496 | No | ||
| 11 | PHCA | 12301 7263 | 1671 | 0.172 | 0.0457 | No | ||
| 12 | RHOG | 17727 | 1773 | 0.160 | 0.0471 | No | ||
| 13 | CLPTM1 | 17940 | 1779 | 0.160 | 0.0536 | No | ||
| 14 | PARD6A | 12142 | 1887 | 0.146 | 0.0540 | No | ||
| 15 | PSMC6 | 7379 | 2078 | 0.124 | 0.0490 | No | ||
| 16 | TMEM24 | 19150 | 2185 | 0.115 | 0.0482 | No | ||
| 17 | CORO1C | 16425 | 2273 | 0.106 | 0.0479 | No | ||
| 18 | RAB35 | 13386 | 2278 | 0.105 | 0.0522 | No | ||
| 19 | LASP1 | 4986 4985 1248 | 2468 | 0.090 | 0.0458 | No | ||
| 20 | TEAD2 | 18252 | 2518 | 0.087 | 0.0469 | No | ||
| 21 | ADAM17 | 4343 | 2563 | 0.084 | 0.0481 | No | ||
| 22 | MYO1C | 20777 | 2631 | 0.080 | 0.0478 | No | ||
| 23 | SEC24C | 22086 | 2699 | 0.075 | 0.0474 | No | ||
| 24 | PIAS1 | 7126 | 2714 | 0.074 | 0.0498 | No | ||
| 25 | AP1GBP1 | 20725 | 2759 | 0.072 | 0.0505 | No | ||
| 26 | EPS15 | 4677 | 2801 | 0.069 | 0.0512 | No | ||
| 27 | KLF5 | 4456 | 2897 | 0.063 | 0.0487 | No | ||
| 28 | FKBP8 | 18587 | 2914 | 0.063 | 0.0506 | No | ||
| 29 | BCL7C | 4442 | 3070 | 0.055 | 0.0445 | No | ||
| 30 | GABARAPL2 | 8211 13552 | 3243 | 0.048 | 0.0372 | No | ||
| 31 | ACE | 1419 4313 | 3300 | 0.046 | 0.0361 | No | ||
| 32 | SNAPAP | 15270 | 3441 | 0.041 | 0.0303 | No | ||
| 33 | IQGAP1 | 6619 | 3500 | 0.039 | 0.0288 | No | ||
| 34 | RELA | 23783 | 3534 | 0.038 | 0.0287 | No | ||
| 35 | VASP | 5847 | 3549 | 0.038 | 0.0295 | No | ||
| 36 | SLC35F5 | 13171 | 3592 | 0.037 | 0.0288 | No | ||
| 37 | MAZ | 1327 17623 | 3641 | 0.035 | 0.0277 | No | ||
| 38 | PLEKHM1 | 20196 1449 | 3680 | 0.034 | 0.0271 | No | ||
| 39 | KLF2 | 9229 | 3686 | 0.034 | 0.0283 | No | ||
| 40 | NFYB | 5173 | 3713 | 0.034 | 0.0283 | No | ||
| 41 | ROM1 | 23751 | 3766 | 0.033 | 0.0269 | No | ||
| 42 | GLTP | 16419 | 3872 | 0.030 | 0.0225 | No | ||
| 43 | XPR1 | 5383 | 3876 | 0.030 | 0.0236 | No | ||
| 44 | SMOC1 | 21221 | 3883 | 0.030 | 0.0246 | No | ||
| 45 | SLC18A3 | 21883 | 4020 | 0.027 | 0.0184 | No | ||
| 46 | ADCK1 | 21191 | 4322 | 0.022 | 0.0030 | No | ||
| 47 | DUOX1 | 14879 | 4324 | 0.022 | 0.0039 | No | ||
| 48 | SMARCD1 | 2241 22360 2250 | 4522 | 0.020 | -0.0060 | No | ||
| 49 | WNT2B | 15214 1838 | 4713 | 0.018 | -0.0155 | No | ||
| 50 | TAGLN | 19136 | 4842 | 0.016 | -0.0218 | No | ||
| 51 | RAB26 | 11609 | 4918 | 0.016 | -0.0251 | No | ||
| 52 | BRI3BP | 16696 | 4998 | 0.015 | -0.0288 | No | ||
| 53 | SUV39H2 | 7232 12263 | 5083 | 0.015 | -0.0327 | No | ||
| 54 | RAB2B | 21842 | 5122 | 0.014 | -0.0342 | No | ||
| 55 | PIAS3 | 15491 672 1906 | 5601 | 0.011 | -0.0596 | No | ||
| 56 | TLX2 | 5773 1165 | 5750 | 0.011 | -0.0671 | No | ||
| 57 | OTX1 | 20517 | 5923 | 0.010 | -0.0760 | No | ||
| 58 | MAPK7 | 1381 20414 | 5971 | 0.010 | -0.0782 | No | ||
| 59 | PVRL2 | 9671 5336 | 6192 | 0.009 | -0.0897 | No | ||
| 60 | JUB | 21834 3339 | 6363 | 0.008 | -0.0986 | No | ||
| 61 | EDG8 | 19212 | 6687 | 0.007 | -0.1158 | No | ||
| 62 | APLP1 | 17890 | 6994 | 0.006 | -0.1321 | No | ||
| 63 | SLC30A3 | 5995 | 7024 | 0.006 | -0.1334 | No | ||
| 64 | FDX1 | 19117 | 7031 | 0.006 | -0.1335 | No | ||
| 65 | ELAVL3 | 9136 | 8118 | 0.003 | -0.1922 | No | ||
| 66 | BCL2L2 | 8653 4441 | 8391 | 0.003 | -0.2069 | No | ||
| 67 | TIMELESS | 10177 | 8688 | 0.002 | -0.2228 | No | ||
| 68 | ELAC2 | 1267 20843 | 9190 | 0.001 | -0.2499 | No | ||
| 69 | BCL9L | 19475 | 9246 | 0.001 | -0.2528 | No | ||
| 70 | IRX3 | 18793 | 9560 | 0.000 | -0.2698 | No | ||
| 71 | PGM2L1 | 7712 | 9613 | 0.000 | -0.2726 | No | ||
| 72 | CRY1 | 19662 | 9653 | 0.000 | -0.2747 | No | ||
| 73 | KCNQ4 | 12247 2533 | 9819 | -0.000 | -0.2836 | No | ||
| 74 | ASCL2 | 5079 | 10044 | -0.001 | -0.2957 | No | ||
| 75 | KTN1 | 2865 22034 2586 | 10091 | -0.001 | -0.2982 | No | ||
| 76 | ACBD5 | 15105 2751 | 10329 | -0.001 | -0.3110 | No | ||
| 77 | NXPH4 | 19602 | 10645 | -0.002 | -0.3280 | No | ||
| 78 | PIP5K2B | 20269 1254 | 10847 | -0.002 | -0.3388 | No | ||
| 79 | SLC24A6 | 16721 | 10970 | -0.002 | -0.3453 | No | ||
| 80 | CACNA1A | 4461 | 10987 | -0.002 | -0.3461 | No | ||
| 81 | HTATIP | 3690 | 11227 | -0.003 | -0.3589 | No | ||
| 82 | NOTCH3 | 23031 | 11303 | -0.003 | -0.3629 | No | ||
| 83 | CSPG4 | 19435 15262 | 11318 | -0.003 | -0.3635 | No | ||
| 84 | CHAT | 21882 | 11371 | -0.003 | -0.3662 | No | ||
| 85 | MAP2K7 | 6453 | 11434 | -0.003 | -0.3694 | No | ||
| 86 | LOXL4 | 23672 | 11590 | -0.004 | -0.3777 | No | ||
| 87 | PIGW | 20312 | 11741 | -0.004 | -0.3856 | No | ||
| 88 | SFRS2 | 9807 20136 | 12005 | -0.004 | -0.3997 | No | ||
| 89 | PTPN2 | 23414 | 12037 | -0.004 | -0.4012 | No | ||
| 90 | CBLN1 | 18800 | 12174 | -0.005 | -0.4083 | No | ||
| 91 | CNNM4 | 14273 | 12181 | -0.005 | -0.4085 | No | ||
| 92 | A2BP1 | 11212 1652 1689 | 12186 | -0.005 | -0.4085 | No | ||
| 93 | FOSB | 17945 | 12838 | -0.007 | -0.4435 | No | ||
| 94 | ABCC1 | 1668 5087 | 13107 | -0.007 | -0.4577 | No | ||
| 95 | HCN4 | 9080 | 13157 | -0.008 | -0.4600 | No | ||
| 96 | HOXC13 | 22344 | 13190 | -0.008 | -0.4614 | No | ||
| 97 | FGF11 | 20383 | 13286 | -0.008 | -0.4662 | No | ||
| 98 | PIGN | 13865 | 13290 | -0.008 | -0.4660 | No | ||
| 99 | HES7 | 20828 | 13355 | -0.008 | -0.4691 | No | ||
| 100 | EFNB3 | 20391 | 13361 | -0.008 | -0.4691 | No | ||
| 101 | IDH3A | 19447 | 13566 | -0.009 | -0.4797 | No | ||
| 102 | PTCH2 | 9640 | 13706 | -0.010 | -0.4868 | No | ||
| 103 | GGN | 2472 1171 18312 | 14042 | -0.011 | -0.5045 | No | ||
| 104 | SLC1A2 | 5453 | 14145 | -0.012 | -0.5095 | No | ||
| 105 | CAMK2G | 21905 | 14433 | -0.014 | -0.5245 | No | ||
| 106 | ANKRD25 | 6161 | 14553 | -0.015 | -0.5303 | No | ||
| 107 | HTR7 | 23690 | 14623 | -0.015 | -0.5334 | No | ||
| 108 | GPR3 | 15729 | 14741 | -0.016 | -0.5390 | No | ||
| 109 | PHOX2A | 18168 | 14759 | -0.016 | -0.5392 | No | ||
| 110 | CYP26B1 | 17093 | 15018 | -0.020 | -0.5524 | No | ||
| 111 | NTF5 | 18248 | 15099 | -0.021 | -0.5558 | No | ||
| 112 | ECE1 | 2427 10516 | 15204 | -0.022 | -0.5605 | No | ||
| 113 | CXCL12 | 9792 1182 1031 | 15675 | -0.033 | -0.5846 | No | ||
| 114 | MAP3K6 | 16054 | 15681 | -0.033 | -0.5834 | No | ||
| 115 | ADAM15 | 15275 | 15688 | -0.033 | -0.5823 | No | ||
| 116 | TCF4 | 5642 | 15778 | -0.037 | -0.5856 | No | ||
| 117 | EFNA3 | 15278 | 15955 | -0.044 | -0.5933 | No | ||
| 118 | CDC37 | 19214 | 15994 | -0.045 | -0.5934 | No | ||
| 119 | GRB2 | 20149 | 16030 | -0.047 | -0.5933 | No | ||
| 120 | EPS8L2 | 18012 | 16036 | -0.047 | -0.5915 | No | ||
| 121 | NAV1 | 3978 5681 | 16192 | -0.055 | -0.5976 | No | ||
| 122 | SPATA6 | 16134 | 16304 | -0.062 | -0.6010 | No | ||
| 123 | CPD | 20344 4556 | 16454 | -0.072 | -0.6060 | No | ||
| 124 | DNAJC4 | 23796 | 16615 | -0.085 | -0.6110 | Yes | ||
| 125 | NET1 | 7100 | 16631 | -0.087 | -0.6081 | Yes | ||
| 126 | CALM3 | 8682 | 16715 | -0.094 | -0.6086 | Yes | ||
| 127 | SHMT1 | 5431 | 16719 | -0.095 | -0.6047 | Yes | ||
| 128 | MYOHD1 | 20724 | 16784 | -0.101 | -0.6039 | Yes | ||
| 129 | POLR3E | 18100 | 16874 | -0.110 | -0.6040 | Yes | ||
| 130 | CSNK1D | 4181 8352 1387 | 16987 | -0.124 | -0.6048 | Yes | ||
| 131 | DRAP1 | 23979 | 17030 | -0.129 | -0.6016 | Yes | ||
| 132 | ELL | 18586 | 17049 | -0.131 | -0.5970 | Yes | ||
| 133 | PER1 | 20827 | 17062 | -0.134 | -0.5919 | Yes | ||
| 134 | GCN5L2 | 4763 | 17086 | -0.137 | -0.5873 | Yes | ||
| 135 | TGFB1 | 18332 | 17111 | -0.141 | -0.5826 | Yes | ||
| 136 | PTOV1 | 13522 | 17137 | -0.144 | -0.5778 | Yes | ||
| 137 | MAP3K11 | 11163 | 17143 | -0.146 | -0.5719 | Yes | ||
| 138 | PEX14 | 15668 | 17145 | -0.146 | -0.5657 | Yes | ||
| 139 | ULK1 | 16436 | 17149 | -0.146 | -0.5597 | Yes | ||
| 140 | SIPA1 | 9820 | 17238 | -0.160 | -0.5576 | Yes | ||
| 141 | SENP1 | 10301 | 17291 | -0.167 | -0.5533 | Yes | ||
| 142 | TRIM28 | 18389 | 17315 | -0.172 | -0.5472 | Yes | ||
| 143 | PITPNM1 | 23770 | 17519 | -0.210 | -0.5493 | Yes | ||
| 144 | TAGLN2 | 14043 | 17746 | -0.256 | -0.5506 | Yes | ||
| 145 | SCYL1 | 23986 | 17819 | -0.277 | -0.5427 | Yes | ||
| 146 | PAK4 | 17909 | 17866 | -0.291 | -0.5328 | Yes | ||
| 147 | SMARCE1 | 1218 20253 | 17880 | -0.298 | -0.5208 | Yes | ||
| 148 | LDB1 | 23654 | 17896 | -0.304 | -0.5087 | Yes | ||
| 149 | RABEP2 | 18082 | 18032 | -0.351 | -0.5010 | Yes | ||
| 150 | UPF2 | 11591 | 18058 | -0.365 | -0.4868 | Yes | ||
| 151 | LZTS2 | 10362 | 18110 | -0.385 | -0.4731 | Yes | ||
| 152 | GOLGA3 | 6535 | 18114 | -0.388 | -0.4568 | Yes | ||
| 153 | VAMP2 | 20826 | 18175 | -0.423 | -0.4420 | Yes | ||
| 154 | MAP4K2 | 6457 | 18228 | -0.460 | -0.4252 | Yes | ||
| 155 | APLP2 | 19190 | 18258 | -0.480 | -0.4063 | Yes | ||
| 156 | ATP2A2 | 4421 3481 | 18345 | -0.567 | -0.3868 | Yes | ||
| 157 | NLK | 5179 5178 | 18353 | -0.578 | -0.3625 | Yes | ||
| 158 | INPPL1 | 17733 3775 | 18372 | -0.598 | -0.3380 | Yes | ||
| 159 | DGKA | 3359 19589 | 18482 | -0.843 | -0.3080 | Yes | ||
| 160 | ANP32A | 8592 4387 19415 | 18574 | -1.725 | -0.2394 | Yes | ||
| 161 | NFAT5 | 3921 7037 12036 | 18604 | -2.808 | -0.1213 | Yes | ||
| 162 | GAS7 | 20836 1299 | 18606 | -2.859 | 0.0005 | Yes |