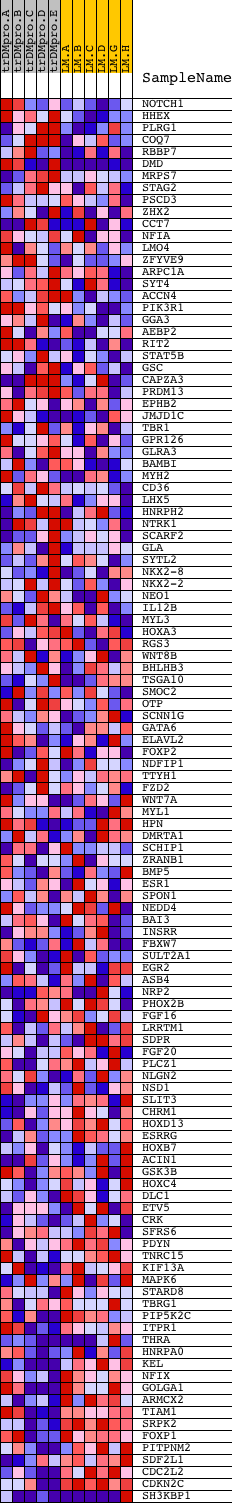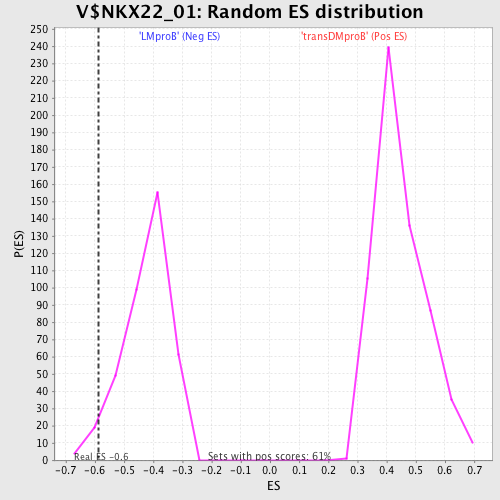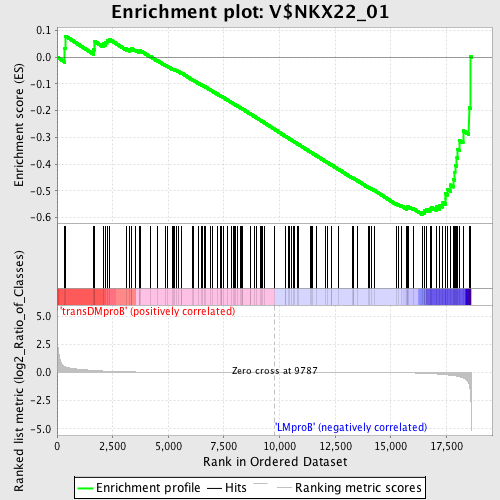
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$NKX22_01 |
| Enrichment Score (ES) | -0.58841026 |
| Normalized Enrichment Score (NES) | -1.3941946 |
| Nominal p-value | 0.03875969 |
| FDR q-value | 0.54113525 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NOTCH1 | 14649 | 347 | 0.525 | 0.0324 | No | ||
| 2 | HHEX | 23872 | 384 | 0.493 | 0.0785 | No | ||
| 3 | PLRG1 | 15565 | 1637 | 0.175 | 0.0279 | No | ||
| 4 | COQ7 | 17664 | 1666 | 0.172 | 0.0432 | No | ||
| 5 | RBBP7 | 10887 6366 10886 | 1667 | 0.172 | 0.0600 | No | ||
| 6 | DMD | 24295 2647 | 2070 | 0.125 | 0.0504 | No | ||
| 7 | MRPS7 | 20595 | 2174 | 0.115 | 0.0561 | No | ||
| 8 | STAG2 | 5521 | 2243 | 0.109 | 0.0630 | No | ||
| 9 | PSCD3 | 16633 | 2344 | 0.100 | 0.0674 | No | ||
| 10 | ZHX2 | 11842 | 3117 | 0.053 | 0.0308 | No | ||
| 11 | CCT7 | 17385 | 3271 | 0.047 | 0.0271 | No | ||
| 12 | NFIA | 16172 5170 | 3326 | 0.045 | 0.0285 | No | ||
| 13 | LMO4 | 15151 | 3331 | 0.045 | 0.0327 | No | ||
| 14 | ZFYVE9 | 10501 2449 | 3515 | 0.039 | 0.0266 | No | ||
| 15 | ARPC1A | 3541 16628 3519 | 3689 | 0.034 | 0.0205 | No | ||
| 16 | SYT4 | 5566 | 3704 | 0.034 | 0.0231 | No | ||
| 17 | ACCN4 | 10795 | 3736 | 0.033 | 0.0247 | No | ||
| 18 | PIK3R1 | 3170 | 4187 | 0.024 | 0.0027 | No | ||
| 19 | GGA3 | 1206 6435 | 4511 | 0.020 | -0.0129 | No | ||
| 20 | AEBP2 | 4359 | 4852 | 0.016 | -0.0296 | No | ||
| 21 | RIT2 | 5380 | 4971 | 0.015 | -0.0345 | No | ||
| 22 | STAT5B | 20222 | 5199 | 0.014 | -0.0454 | No | ||
| 23 | GSC | 20990 | 5247 | 0.013 | -0.0467 | No | ||
| 24 | CAPZA3 | 17252 | 5283 | 0.013 | -0.0473 | No | ||
| 25 | PRDM13 | 15933 | 5358 | 0.013 | -0.0500 | No | ||
| 26 | EPHB2 | 4675 2440 8910 | 5459 | 0.012 | -0.0543 | No | ||
| 27 | JMJD1C | 11531 19996 | 5574 | 0.012 | -0.0593 | No | ||
| 28 | TBR1 | 83 | 5604 | 0.011 | -0.0598 | No | ||
| 29 | GPR126 | 19816 | 6078 | 0.009 | -0.0844 | No | ||
| 30 | GLRA3 | 18617 | 6119 | 0.009 | -0.0857 | No | ||
| 31 | BAMBI | 23629 | 6341 | 0.008 | -0.0969 | No | ||
| 32 | MYH2 | 20838 | 6486 | 0.008 | -0.1039 | No | ||
| 33 | CD36 | 8712 | 6537 | 0.007 | -0.1059 | No | ||
| 34 | LHX5 | 9274 | 6608 | 0.007 | -0.1090 | No | ||
| 35 | HNRPH2 | 12085 7082 | 6647 | 0.007 | -0.1103 | No | ||
| 36 | NTRK1 | 15299 | 6881 | 0.006 | -0.1223 | No | ||
| 37 | SCARF2 | 22833 | 6998 | 0.006 | -0.1280 | No | ||
| 38 | GLA | 4362 | 7187 | 0.006 | -0.1376 | No | ||
| 39 | SYTL2 | 18190 1539 3730 | 7347 | 0.005 | -0.1457 | No | ||
| 40 | NKX2-8 | 9465 | 7386 | 0.005 | -0.1473 | No | ||
| 41 | NKX2-2 | 5176 | 7396 | 0.005 | -0.1473 | No | ||
| 42 | NEO1 | 5161 | 7496 | 0.005 | -0.1522 | No | ||
| 43 | IL12B | 20918 | 7650 | 0.004 | -0.1600 | No | ||
| 44 | MYL3 | 19291 | 7837 | 0.004 | -0.1697 | No | ||
| 45 | HOXA3 | 9108 1075 | 7917 | 0.004 | -0.1736 | No | ||
| 46 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 7971 | 0.003 | -0.1761 | No | ||
| 47 | WNT8B | 23841 | 8039 | 0.003 | -0.1794 | No | ||
| 48 | BHLHB3 | 16935 | 8117 | 0.003 | -0.1833 | No | ||
| 49 | TSGA10 | 4123 13975 5587 | 8235 | 0.003 | -0.1893 | No | ||
| 50 | SMOC2 | 23378 | 8293 | 0.003 | -0.1921 | No | ||
| 51 | OTP | 21582 | 8333 | 0.003 | -0.1940 | No | ||
| 52 | SCNN1G | 18095 | 8337 | 0.003 | -0.1938 | No | ||
| 53 | GATA6 | 96 | 8682 | 0.002 | -0.2122 | No | ||
| 54 | ELAVL2 | 15839 | 8687 | 0.002 | -0.2123 | No | ||
| 55 | FOXP2 | 4312 17523 8523 | 8879 | 0.002 | -0.2224 | No | ||
| 56 | NDFIP1 | 23573 | 8975 | 0.002 | -0.2274 | No | ||
| 57 | TTYH1 | 2496 7176 | 9143 | 0.001 | -0.2363 | No | ||
| 58 | FZD2 | 12179 | 9189 | 0.001 | -0.2386 | No | ||
| 59 | WNT7A | 17068 | 9215 | 0.001 | -0.2399 | No | ||
| 60 | MYL1 | 4015 | 9319 | 0.001 | -0.2453 | No | ||
| 61 | HPN | 17873 4087 | 9768 | 0.000 | -0.2696 | No | ||
| 62 | DMRTA1 | 16177 | 10259 | -0.001 | -0.2960 | No | ||
| 63 | SCHIP1 | 15569 | 10260 | -0.001 | -0.2959 | No | ||
| 64 | ZRANB1 | 6884 11702 | 10382 | -0.001 | -0.3023 | No | ||
| 65 | BMP5 | 19376 | 10461 | -0.001 | -0.3064 | No | ||
| 66 | ESR1 | 20097 4685 | 10530 | -0.001 | -0.3099 | No | ||
| 67 | SPON1 | 10600 | 10634 | -0.002 | -0.3153 | No | ||
| 68 | NEDD4 | 19384 3101 | 10684 | -0.002 | -0.3178 | No | ||
| 69 | BAI3 | 5580 | 10795 | -0.002 | -0.3236 | No | ||
| 70 | INSRR | 15557 | 10838 | -0.002 | -0.3257 | No | ||
| 71 | FBXW7 | 1805 11928 | 11408 | -0.003 | -0.3561 | No | ||
| 72 | SULT2A1 | 5527 | 11444 | -0.003 | -0.3577 | No | ||
| 73 | EGR2 | 8886 | 11481 | -0.003 | -0.3593 | No | ||
| 74 | ASB4 | 990 17529 | 11678 | -0.004 | -0.3696 | No | ||
| 75 | NRP2 | 5194 9486 5195 | 12077 | -0.005 | -0.3906 | No | ||
| 76 | PHOX2B | 16524 | 12133 | -0.005 | -0.3931 | No | ||
| 77 | FGF16 | 24270 | 12311 | -0.005 | -0.4022 | No | ||
| 78 | LRRTM1 | 17408 | 12345 | -0.005 | -0.4035 | No | ||
| 79 | SDPR | 14252 | 12651 | -0.006 | -0.4194 | No | ||
| 80 | FGF20 | 18892 | 13281 | -0.008 | -0.4526 | No | ||
| 81 | PLCZ1 | 16945 | 13299 | -0.008 | -0.4527 | No | ||
| 82 | NLGN2 | 10117 | 13300 | -0.008 | -0.4519 | No | ||
| 83 | NSD1 | 2134 5197 | 13515 | -0.009 | -0.4626 | No | ||
| 84 | SLIT3 | 20925 | 14010 | -0.011 | -0.4882 | No | ||
| 85 | CHRM1 | 23943 | 14019 | -0.011 | -0.4875 | No | ||
| 86 | HOXD13 | 9114 | 14129 | -0.012 | -0.4923 | No | ||
| 87 | ESRRG | 6447 | 14246 | -0.013 | -0.4973 | No | ||
| 88 | HOXB7 | 666 9110 | 15242 | -0.023 | -0.5489 | No | ||
| 89 | ACIN1 | 3319 2807 21832 | 15325 | -0.025 | -0.5509 | No | ||
| 90 | GSK3B | 22761 | 15479 | -0.028 | -0.5565 | No | ||
| 91 | HOXC4 | 4864 | 15715 | -0.035 | -0.5658 | No | ||
| 92 | DLC1 | 11931 3855 | 15737 | -0.035 | -0.5635 | No | ||
| 93 | ETV5 | 22630 | 15738 | -0.035 | -0.5600 | No | ||
| 94 | CRK | 4559 1249 | 15785 | -0.037 | -0.5589 | No | ||
| 95 | SFRS6 | 14751 | 15999 | -0.046 | -0.5660 | No | ||
| 96 | PDYN | 14429 | 16415 | -0.070 | -0.5816 | Yes | ||
| 97 | TNRC15 | 5971 | 16508 | -0.076 | -0.5792 | Yes | ||
| 98 | KIF13A | 21475 | 16516 | -0.077 | -0.5721 | Yes | ||
| 99 | MAPK6 | 19062 | 16594 | -0.083 | -0.5682 | Yes | ||
| 100 | STARD8 | 10682 | 16762 | -0.099 | -0.5675 | Yes | ||
| 101 | TBRG1 | 3119 19174 | 16831 | -0.105 | -0.5610 | Yes | ||
| 102 | PIP5K2C | 19606 | 17044 | -0.131 | -0.5597 | Yes | ||
| 103 | ITPR1 | 17341 | 17203 | -0.154 | -0.5532 | Yes | ||
| 104 | THRA | 1447 10171 1406 | 17340 | -0.177 | -0.5433 | Yes | ||
| 105 | HNRPA0 | 13382 | 17459 | -0.199 | -0.5303 | Yes | ||
| 106 | KEL | 17170 | 17461 | -0.199 | -0.5110 | Yes | ||
| 107 | NFIX | 9458 | 17551 | -0.217 | -0.4947 | Yes | ||
| 108 | GOLGA1 | 14595 | 17681 | -0.243 | -0.4780 | Yes | ||
| 109 | ARMCX2 | 12530 | 17826 | -0.279 | -0.4586 | Yes | ||
| 110 | TIAM1 | 22547 | 17878 | -0.297 | -0.4324 | Yes | ||
| 111 | SRPK2 | 5513 | 17885 | -0.299 | -0.4037 | Yes | ||
| 112 | FOXP1 | 4242 | 17971 | -0.329 | -0.3762 | Yes | ||
| 113 | PITPNM2 | 16371 | 18012 | -0.345 | -0.3447 | Yes | ||
| 114 | SDF2L1 | 22649 | 18085 | -0.375 | -0.3120 | Yes | ||
| 115 | CDC2L2 | 15964 2419 | 18247 | -0.473 | -0.2746 | Yes | ||
| 116 | CDKN2C | 15806 2321 | 18513 | -1.023 | -0.1892 | Yes | ||
| 117 | SH3KBP1 | 2631 12206 7189 | 18578 | -1.998 | 0.0021 | Yes |