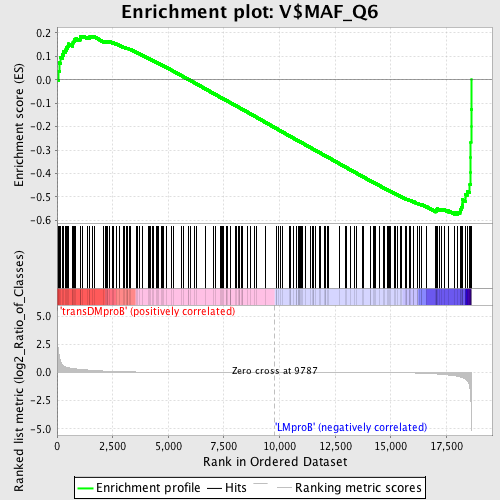
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$MAF_Q6 |
| Enrichment Score (ES) | -0.575123 |
| Normalized Enrichment Score (NES) | -1.4450018 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.4934762 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FGF13 | 4720 8963 2627 | 81 | 1.558 | 0.0351 | No | ||
| 2 | CD79B | 20185 1309 | 99 | 1.495 | 0.0721 | No | ||
| 3 | LAMP2 | 9267 2653 | 162 | 0.982 | 0.0936 | No | ||
| 4 | HMGN2 | 9095 | 238 | 0.678 | 0.1067 | No | ||
| 5 | SQLE | 22468 2229 | 282 | 0.592 | 0.1194 | No | ||
| 6 | RASGRP3 | 10767 | 375 | 0.500 | 0.1271 | No | ||
| 7 | CHIC2 | 16510 | 409 | 0.466 | 0.1371 | No | ||
| 8 | PIK3CG | 6635 | 484 | 0.423 | 0.1438 | No | ||
| 9 | ALDOA | 8572 | 509 | 0.407 | 0.1529 | No | ||
| 10 | RASSF3 | 19616 | 709 | 0.341 | 0.1507 | No | ||
| 11 | LMAN2 | 21454 | 712 | 0.340 | 0.1592 | No | ||
| 12 | BAD | 24000 | 757 | 0.325 | 0.1651 | No | ||
| 13 | DYRK1A | 4649 | 794 | 0.317 | 0.1711 | No | ||
| 14 | VCAM1 | 5851 | 846 | 0.306 | 0.1761 | No | ||
| 15 | RRAS | 18256 | 1029 | 0.273 | 0.1732 | No | ||
| 16 | HMGCS1 | 9917 | 1044 | 0.269 | 0.1792 | No | ||
| 17 | CBX8 | 11401 | 1047 | 0.268 | 0.1859 | No | ||
| 18 | AP2M1 | 8603 | 1158 | 0.248 | 0.1863 | No | ||
| 19 | PGF | 21020 | 1354 | 0.217 | 0.1812 | No | ||
| 20 | BMP4 | 21863 | 1463 | 0.201 | 0.1804 | No | ||
| 21 | BRDT | 4327 | 1473 | 0.200 | 0.1850 | No | ||
| 22 | VAMP3 | 15660 | 1594 | 0.181 | 0.1831 | No | ||
| 23 | STK4 | 14743 | 1661 | 0.173 | 0.1839 | No | ||
| 24 | AKT3 | 13739 982 | 2091 | 0.122 | 0.1637 | No | ||
| 25 | ETV3 | 11294 | 2160 | 0.117 | 0.1630 | No | ||
| 26 | HMGA1 | 23323 | 2232 | 0.111 | 0.1619 | No | ||
| 27 | CORO1C | 16425 | 2273 | 0.106 | 0.1624 | No | ||
| 28 | CLNS1A | 4526 4525 | 2285 | 0.105 | 0.1645 | No | ||
| 29 | SLC9A3R1 | 11250 | 2362 | 0.098 | 0.1628 | No | ||
| 30 | BMF | 5074 | 2502 | 0.088 | 0.1575 | No | ||
| 31 | EDG2 | 9033 | 2525 | 0.087 | 0.1585 | No | ||
| 32 | PTGER2 | 22039 | 2668 | 0.077 | 0.1528 | No | ||
| 33 | EPS15 | 4677 | 2801 | 0.069 | 0.1474 | No | ||
| 34 | AGPAT4 | 23384 | 2978 | 0.060 | 0.1393 | No | ||
| 35 | SATB1 | 5408 | 3048 | 0.056 | 0.1370 | No | ||
| 36 | ZHX2 | 11842 | 3117 | 0.053 | 0.1346 | No | ||
| 37 | CPNE1 | 11186 | 3146 | 0.052 | 0.1344 | No | ||
| 38 | GPX1 | 19310 | 3178 | 0.050 | 0.1340 | No | ||
| 39 | SYNCRIP | 3078 3035 3107 | 3268 | 0.047 | 0.1304 | No | ||
| 40 | UNC5C | 10256 | 3302 | 0.046 | 0.1297 | No | ||
| 41 | ALCAM | 4367 | 3573 | 0.037 | 0.1160 | No | ||
| 42 | RAB33A | 358 | 3624 | 0.036 | 0.1142 | No | ||
| 43 | POU2AF1 | 19455 | 3699 | 0.034 | 0.1111 | No | ||
| 44 | AIF1 | 1582 23005 | 3846 | 0.031 | 0.1039 | No | ||
| 45 | CHD2 | 10847 | 4086 | 0.026 | 0.0916 | No | ||
| 46 | WNT3 | 20635 | 4150 | 0.025 | 0.0888 | No | ||
| 47 | SLA | 5449 | 4189 | 0.024 | 0.0874 | No | ||
| 48 | PRDX6 | 4389 8596 | 4289 | 0.022 | 0.0826 | No | ||
| 49 | SPAG9 | 1322 12904 7707 | 4343 | 0.022 | 0.0802 | No | ||
| 50 | CNTN4 | 11265 1088 17345 | 4478 | 0.020 | 0.0735 | No | ||
| 51 | STAC2 | 20265 | 4529 | 0.020 | 0.0713 | No | ||
| 52 | PROK2 | 17057 | 4560 | 0.019 | 0.0701 | No | ||
| 53 | HOXC6 | 22339 2302 | 4688 | 0.018 | 0.0637 | No | ||
| 54 | CD5 | 23741 | 4752 | 0.017 | 0.0607 | No | ||
| 55 | KCTD15 | 17864 | 4801 | 0.017 | 0.0585 | No | ||
| 56 | TWIST1 | 21291 | 4910 | 0.016 | 0.0531 | No | ||
| 57 | NUMB | 5202 2146 2054 | 5132 | 0.014 | 0.0414 | No | ||
| 58 | MITF | 17349 | 5215 | 0.014 | 0.0373 | No | ||
| 59 | JMJD1C | 11531 19996 | 5574 | 0.012 | 0.0182 | No | ||
| 60 | BNC2 | 15850 | 5694 | 0.011 | 0.0120 | No | ||
| 61 | HAS2 | 22293 | 5921 | 0.010 | 0.0000 | No | ||
| 62 | OTX1 | 20517 | 5923 | 0.010 | 0.0002 | No | ||
| 63 | ITPKB | 3963 14028 | 5984 | 0.010 | -0.0028 | No | ||
| 64 | PIK4CB | 1875 15511 | 6184 | 0.009 | -0.0134 | No | ||
| 65 | RPS6KA3 | 8490 | 6261 | 0.008 | -0.0173 | No | ||
| 66 | NPAS3 | 2077 21273 | 6648 | 0.007 | -0.0381 | No | ||
| 67 | TPBG | 5793 | 7014 | 0.006 | -0.0577 | No | ||
| 68 | NRG2 | 4274 | 7132 | 0.006 | -0.0639 | No | ||
| 69 | TFEB | 23207 | 7337 | 0.005 | -0.0749 | No | ||
| 70 | NKX2-2 | 5176 | 7396 | 0.005 | -0.0779 | No | ||
| 71 | SNCG | 21878 | 7435 | 0.005 | -0.0798 | No | ||
| 72 | HMGB3 | 9096 | 7483 | 0.005 | -0.0823 | No | ||
| 73 | PDGFRA | 16824 | 7618 | 0.004 | -0.0894 | No | ||
| 74 | STX12 | 4140 | 7628 | 0.004 | -0.0898 | No | ||
| 75 | MEOX2 | 21285 | 7638 | 0.004 | -0.0902 | No | ||
| 76 | CHAD | 20702 | 7785 | 0.004 | -0.0980 | No | ||
| 77 | SOX2 | 9849 15612 | 7808 | 0.004 | -0.0991 | No | ||
| 78 | TLX1 | 23839 | 7811 | 0.004 | -0.0991 | No | ||
| 79 | GPR82 | 24376 | 8003 | 0.003 | -0.1094 | No | ||
| 80 | ESM1 | 21560 | 8055 | 0.003 | -0.1121 | No | ||
| 81 | CBLN2 | 23498 | 8161 | 0.003 | -0.1177 | No | ||
| 82 | PNRC1 | 15925 | 8191 | 0.003 | -0.1192 | No | ||
| 83 | KCNJ16 | 20613 | 8218 | 0.003 | -0.1205 | No | ||
| 84 | LALBA | 22141 | 8306 | 0.003 | -0.1252 | No | ||
| 85 | ACTR2 | 20523 | 8321 | 0.003 | -0.1258 | No | ||
| 86 | OTP | 21582 | 8333 | 0.003 | -0.1264 | No | ||
| 87 | ADORA3 | 4352 | 8550 | 0.002 | -0.1380 | No | ||
| 88 | TIMELESS | 10177 | 8688 | 0.002 | -0.1454 | No | ||
| 89 | FOXP2 | 4312 17523 8523 | 8879 | 0.002 | -0.1557 | No | ||
| 90 | SLC41A1 | 8269 | 8979 | 0.002 | -0.1610 | No | ||
| 91 | ROD1 | 6047 2317 | 9366 | 0.001 | -0.1819 | No | ||
| 92 | SP3 | 5483 | 9875 | -0.000 | -0.2095 | No | ||
| 93 | SIX4 | 5445 | 9876 | -0.000 | -0.2095 | No | ||
| 94 | PCDH10 | 5227 9534 1893 | 9958 | -0.000 | -0.2139 | No | ||
| 95 | AMOT | 11341 24037 6587 | 10021 | -0.000 | -0.2172 | No | ||
| 96 | ALX3 | 15456 | 10118 | -0.001 | -0.2224 | No | ||
| 97 | BMP5 | 19376 | 10461 | -0.001 | -0.2409 | No | ||
| 98 | HOXB8 | 9111 | 10475 | -0.001 | -0.2416 | No | ||
| 99 | TUSC3 | 18629 | 10488 | -0.001 | -0.2422 | No | ||
| 100 | NXPH4 | 19602 | 10645 | -0.002 | -0.2507 | No | ||
| 101 | SYNPR | 22094 | 10752 | -0.002 | -0.2564 | No | ||
| 102 | NCOA3 | 5156 | 10763 | -0.002 | -0.2569 | No | ||
| 103 | KDELR2 | 16634 12428 | 10834 | -0.002 | -0.2606 | No | ||
| 104 | INSRR | 15557 | 10838 | -0.002 | -0.2607 | No | ||
| 105 | RARA | 5358 | 10889 | -0.002 | -0.2634 | No | ||
| 106 | FXYD4 | 17030 1047 | 10895 | -0.002 | -0.2636 | No | ||
| 107 | GRM3 | 16929 | 10957 | -0.002 | -0.2669 | No | ||
| 108 | SMARCA1 | 24165 | 10996 | -0.002 | -0.2689 | No | ||
| 109 | CDH13 | 4506 3826 | 11043 | -0.002 | -0.2713 | No | ||
| 110 | TRERF1 | 1546 23213 | 11183 | -0.003 | -0.2788 | No | ||
| 111 | CHM | 4521 | 11373 | -0.003 | -0.2890 | No | ||
| 112 | TITF1 | 10180 21062 | 11473 | -0.003 | -0.2942 | No | ||
| 113 | RARB | 3011 21915 | 11529 | -0.003 | -0.2971 | No | ||
| 114 | CUL2 | 23630 | 11532 | -0.003 | -0.2972 | No | ||
| 115 | TGFBR1 | 16213 10165 5745 | 11612 | -0.004 | -0.3014 | No | ||
| 116 | KPNB1 | 20274 | 11790 | -0.004 | -0.3109 | No | ||
| 117 | PAX6 | 5223 | 11820 | -0.004 | -0.3123 | No | ||
| 118 | SIX1 | 9821 | 12004 | -0.004 | -0.3222 | No | ||
| 119 | COL1A1 | 8771 | 12043 | -0.005 | -0.3241 | No | ||
| 120 | CXXC5 | 12523 | 12063 | -0.005 | -0.3250 | No | ||
| 121 | SNAP25 | 5466 | 12140 | -0.005 | -0.3290 | No | ||
| 122 | A2BP1 | 11212 1652 1689 | 12186 | -0.005 | -0.3313 | No | ||
| 123 | STAT6 | 19854 9909 | 12671 | -0.006 | -0.3574 | No | ||
| 124 | SLC34A3 | 14664 | 12945 | -0.007 | -0.3721 | No | ||
| 125 | ADAM2 | 21777 | 13001 | -0.007 | -0.3749 | No | ||
| 126 | FLOT2 | 1247 8975 1430 | 13169 | -0.008 | -0.3838 | No | ||
| 127 | EFNB3 | 20391 | 13361 | -0.008 | -0.3939 | No | ||
| 128 | GAB2 | 1821 18184 2025 | 13442 | -0.009 | -0.3980 | No | ||
| 129 | TCF2 | 1395 20727 | 13705 | -0.010 | -0.4120 | No | ||
| 130 | RORA | 3019 5388 | 13769 | -0.010 | -0.4152 | No | ||
| 131 | CREB3L2 | 17182 | 14071 | -0.012 | -0.4312 | No | ||
| 132 | GPC4 | 24153 | 14100 | -0.012 | -0.4324 | No | ||
| 133 | VAV2 | 5848 2670 | 14237 | -0.013 | -0.4395 | No | ||
| 134 | ESRRG | 6447 | 14246 | -0.013 | -0.4396 | No | ||
| 135 | NEGR1 | 6806 11572 | 14292 | -0.013 | -0.4417 | No | ||
| 136 | DMP1 | 16776 | 14479 | -0.014 | -0.4514 | No | ||
| 137 | FLT1 | 3483 16287 | 14676 | -0.016 | -0.4617 | No | ||
| 138 | MAP4K5 | 11853 21048 2101 2142 | 14705 | -0.016 | -0.4628 | No | ||
| 139 | CALN1 | 4713 | 14845 | -0.017 | -0.4699 | No | ||
| 140 | CACNB3 | 22373 8679 | 14881 | -0.018 | -0.4713 | No | ||
| 141 | SP8 | 21122 | 14946 | -0.019 | -0.4743 | No | ||
| 142 | OGG1 | 17331 | 14997 | -0.019 | -0.4766 | No | ||
| 143 | CNTNAP1 | 11987 | 15171 | -0.022 | -0.4854 | No | ||
| 144 | ELAVL4 | 15805 4889 9137 | 15191 | -0.022 | -0.4859 | No | ||
| 145 | MMRN2 | 4194 | 15285 | -0.024 | -0.4903 | No | ||
| 146 | PPP2R5C | 6522 2147 | 15437 | -0.027 | -0.4978 | No | ||
| 147 | TLK2 | 20628 | 15496 | -0.028 | -0.5002 | No | ||
| 148 | MGLL | 17368 1145 | 15650 | -0.032 | -0.5077 | No | ||
| 149 | SLC35B3 | 3225 21487 | 15693 | -0.033 | -0.5092 | No | ||
| 150 | HOXC4 | 4864 | 15715 | -0.035 | -0.5094 | No | ||
| 151 | GABRA3 | 24139 | 15832 | -0.039 | -0.5147 | No | ||
| 152 | LIN28 | 15723 | 15852 | -0.039 | -0.5148 | No | ||
| 153 | PHF12 | 11192 6483 | 15889 | -0.041 | -0.5157 | No | ||
| 154 | SUPT16H | 8541 | 16016 | -0.046 | -0.5213 | No | ||
| 155 | PTPN7 | 11500 | 16031 | -0.047 | -0.5209 | No | ||
| 156 | POLG | 17789 | 16178 | -0.054 | -0.5275 | No | ||
| 157 | EDG5 | 19216 | 16198 | -0.056 | -0.5271 | No | ||
| 158 | TYSND1 | 20007 | 16289 | -0.061 | -0.5304 | No | ||
| 159 | TTC3 | 1711 5804 1636 1696 1742 1688 10228 1709 1743 | 16391 | -0.068 | -0.5342 | No | ||
| 160 | CDK5 | 16591 | 16392 | -0.068 | -0.5325 | No | ||
| 161 | CDK8 | 6455 6456 3585 3542 | 16607 | -0.084 | -0.5419 | No | ||
| 162 | TNFSF10 | 15626 | 17025 | -0.128 | -0.5613 | No | ||
| 163 | TNKS | 5786 | 17046 | -0.131 | -0.5591 | No | ||
| 164 | HYAL2 | 4891 | 17061 | -0.133 | -0.5565 | No | ||
| 165 | FGD2 | 23309 | 17066 | -0.134 | -0.5533 | No | ||
| 166 | SLC16A6 | 4186 | 17079 | -0.136 | -0.5505 | No | ||
| 167 | ITPR1 | 17341 | 17203 | -0.154 | -0.5532 | No | ||
| 168 | PRKAG1 | 22135 | 17274 | -0.165 | -0.5528 | No | ||
| 169 | HOXB4 | 20686 | 17392 | -0.185 | -0.5545 | No | ||
| 170 | PIK3CD | 9563 | 17581 | -0.222 | -0.5591 | No | ||
| 171 | TIAM1 | 22547 | 17878 | -0.297 | -0.5676 | Yes | ||
| 172 | PHC1 | 17013 | 17998 | -0.341 | -0.5654 | Yes | ||
| 173 | LZTS2 | 10362 | 18110 | -0.385 | -0.5617 | Yes | ||
| 174 | SLC4A2 | 9828 | 18130 | -0.398 | -0.5526 | Yes | ||
| 175 | SYTL1 | 15728 | 18180 | -0.426 | -0.5445 | Yes | ||
| 176 | RNF44 | 8364 | 18204 | -0.441 | -0.5345 | Yes | ||
| 177 | ELOVL6 | 5009 | 18227 | -0.458 | -0.5241 | Yes | ||
| 178 | MAP4K2 | 6457 | 18228 | -0.460 | -0.5125 | Yes | ||
| 179 | UTX | 10266 2574 | 18352 | -0.577 | -0.5045 | Yes | ||
| 180 | NLK | 5179 5178 | 18353 | -0.578 | -0.4899 | Yes | ||
| 181 | SEMA4A | 15289 | 18431 | -0.706 | -0.4762 | Yes | ||
| 182 | CNOT7 | 3787 9600 | 18554 | -1.420 | -0.4468 | Yes | ||
| 183 | PTPN22 | 1775 15470 | 18591 | -2.121 | -0.3949 | Yes | ||
| 184 | DDX50 | 3324 19749 | 18595 | -2.455 | -0.3329 | Yes | ||
| 185 | MPO | 5116 20710 1324 | 18599 | -2.546 | -0.2685 | Yes | ||
| 186 | NFAT5 | 3921 7037 12036 | 18604 | -2.808 | -0.1975 | Yes | ||
| 187 | GAS7 | 20836 1299 | 18606 | -2.859 | -0.1251 | Yes | ||
| 188 | CDC5L | 7746 7745 7744 | 18615 | -4.954 | 0.0001 | Yes |

