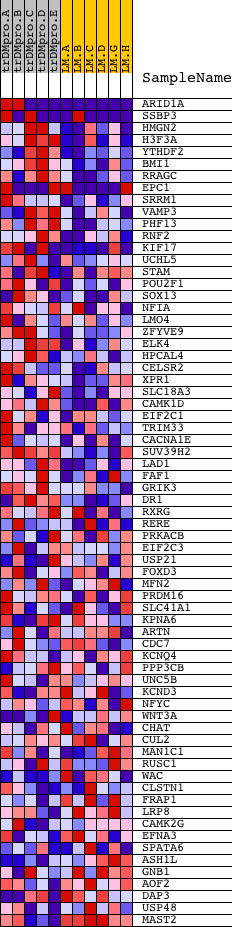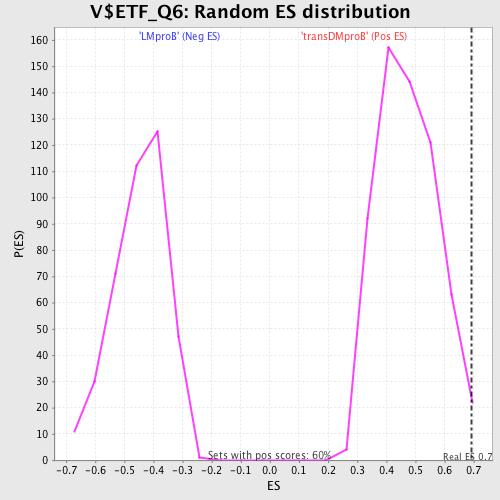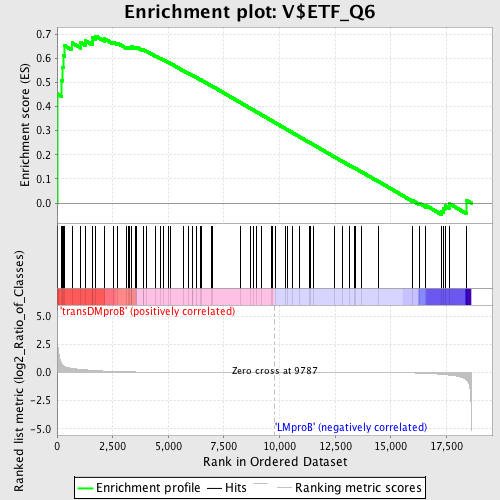
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | V$ETF_Q6 |
| Enrichment Score (ES) | 0.69156003 |
| Normalized Enrichment Score (NES) | 1.4572976 |
| Nominal p-value | 0.013266998 |
| FDR q-value | 0.47933263 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 1 | 5.436 | 0.4527 | Yes | ||
| 2 | SSBP3 | 2439 7814 2431 | 198 | 0.788 | 0.5077 | Yes | ||
| 3 | HMGN2 | 9095 | 238 | 0.678 | 0.5621 | Yes | ||
| 4 | H3F3A | 4838 | 277 | 0.613 | 0.6111 | Yes | ||
| 5 | YTHDF2 | 5623 5624 | 340 | 0.532 | 0.6521 | Yes | ||
| 6 | BMI1 | 4448 15113 | 669 | 0.349 | 0.6634 | Yes | ||
| 7 | RRAGC | 12015 | 1066 | 0.265 | 0.6642 | Yes | ||
| 8 | EPC1 | 2039 8907 2033 | 1268 | 0.229 | 0.6724 | Yes | ||
| 9 | SRRM1 | 6958 | 1578 | 0.183 | 0.6710 | Yes | ||
| 10 | VAMP3 | 15660 | 1594 | 0.181 | 0.6853 | Yes | ||
| 11 | PHF13 | 15659 | 1733 | 0.165 | 0.6916 | Yes | ||
| 12 | RNF2 | 9728 | 2113 | 0.121 | 0.6812 | No | ||
| 13 | KIF17 | 9215 2392 | 2524 | 0.087 | 0.6663 | No | ||
| 14 | UCHL5 | 4108 7076 | 2730 | 0.073 | 0.6614 | No | ||
| 15 | STAM | 2912 15117 | 3118 | 0.053 | 0.6449 | No | ||
| 16 | POU2F1 | 5275 3989 4065 4010 | 3188 | 0.050 | 0.6453 | No | ||
| 17 | SOX13 | 13835 | 3246 | 0.048 | 0.6462 | No | ||
| 18 | NFIA | 16172 5170 | 3326 | 0.045 | 0.6457 | No | ||
| 19 | LMO4 | 15151 | 3331 | 0.045 | 0.6492 | No | ||
| 20 | ZFYVE9 | 10501 2449 | 3515 | 0.039 | 0.6426 | No | ||
| 21 | ELK4 | 8895 4664 3937 | 3547 | 0.038 | 0.6441 | No | ||
| 22 | HPCAL4 | 16096 | 3570 | 0.037 | 0.6460 | No | ||
| 23 | CELSR2 | 15194 12000 | 3869 | 0.030 | 0.6325 | No | ||
| 24 | XPR1 | 5383 | 3876 | 0.030 | 0.6347 | No | ||
| 25 | SLC18A3 | 21883 | 4020 | 0.027 | 0.6292 | No | ||
| 26 | CAMK1D | 5977 2948 | 4430 | 0.021 | 0.6089 | No | ||
| 27 | EIF2C1 | 10672 | 4651 | 0.018 | 0.5985 | No | ||
| 28 | TRIM33 | 8236 13579 15472 8237 | 4774 | 0.017 | 0.5934 | No | ||
| 29 | CACNA1E | 4465 | 5009 | 0.015 | 0.5820 | No | ||
| 30 | SUV39H2 | 7232 12263 | 5083 | 0.015 | 0.5793 | No | ||
| 31 | LAD1 | 14117 | 5667 | 0.011 | 0.5488 | No | ||
| 32 | FAF1 | 2508 16137 8951 | 5889 | 0.010 | 0.5377 | No | ||
| 33 | GRIK3 | 16083 | 6067 | 0.009 | 0.5289 | No | ||
| 34 | DR1 | 16762 | 6106 | 0.009 | 0.5276 | No | ||
| 35 | RXRG | 14063 | 6283 | 0.008 | 0.5188 | No | ||
| 36 | RERE | 12722 | 6454 | 0.008 | 0.5103 | No | ||
| 37 | PRKACB | 15140 | 6469 | 0.008 | 0.5102 | No | ||
| 38 | EIF2C3 | 15757 5645 2318 | 6953 | 0.006 | 0.4847 | No | ||
| 39 | USP21 | 13762 | 6978 | 0.006 | 0.4839 | No | ||
| 40 | FOXD3 | 9088 | 8237 | 0.003 | 0.4163 | No | ||
| 41 | MFN2 | 15678 2417 | 8671 | 0.002 | 0.3931 | No | ||
| 42 | PRDM16 | 12893 | 8812 | 0.002 | 0.3857 | No | ||
| 43 | SLC41A1 | 8269 | 8979 | 0.002 | 0.3769 | No | ||
| 44 | KPNA6 | 4972 | 9199 | 0.001 | 0.3652 | No | ||
| 45 | ARTN | 15783 2430 2532 | 9622 | 0.000 | 0.3425 | No | ||
| 46 | CDC7 | 16765 3477 | 9671 | 0.000 | 0.3399 | No | ||
| 47 | KCNQ4 | 12247 2533 | 9819 | -0.000 | 0.3320 | No | ||
| 48 | PPP3CB | 5285 | 10274 | -0.001 | 0.3076 | No | ||
| 49 | UNC5B | 8404 | 10356 | -0.001 | 0.3033 | No | ||
| 50 | KCND3 | 15464 | 10573 | -0.001 | 0.2918 | No | ||
| 51 | NFYC | 9460 | 10890 | -0.002 | 0.2749 | No | ||
| 52 | WNT3A | 20431 | 11322 | -0.003 | 0.2519 | No | ||
| 53 | CHAT | 21882 | 11371 | -0.003 | 0.2496 | No | ||
| 54 | CUL2 | 23630 | 11532 | -0.003 | 0.2412 | No | ||
| 55 | MAN1C1 | 10514 6064 15714 | 12478 | -0.006 | 0.1908 | No | ||
| 56 | RUSC1 | 1853 15283 | 12808 | -0.007 | 0.1736 | No | ||
| 57 | WAC | 5905 5906 | 13124 | -0.008 | 0.1572 | No | ||
| 58 | CLSTN1 | 7241 | 13344 | -0.008 | 0.1461 | No | ||
| 59 | FRAP1 | 2468 15991 | 13428 | -0.009 | 0.1423 | No | ||
| 60 | LRP8 | 16149 2425 | 13687 | -0.010 | 0.1292 | No | ||
| 61 | CAMK2G | 21905 | 14433 | -0.014 | 0.0902 | No | ||
| 62 | EFNA3 | 15278 | 15955 | -0.044 | 0.0118 | No | ||
| 63 | SPATA6 | 16134 | 16304 | -0.062 | -0.0018 | No | ||
| 64 | ASH1L | 5311 15546 15547 | 16578 | -0.082 | -0.0097 | No | ||
| 65 | GNB1 | 15967 | 17272 | -0.165 | -0.0333 | No | ||
| 66 | AOF2 | 13625 | 17359 | -0.180 | -0.0230 | No | ||
| 67 | DAP3 | 7238 12271 1811 | 17436 | -0.193 | -0.0110 | No | ||
| 68 | USP48 | 16022 | 17619 | -0.230 | -0.0017 | No | ||
| 69 | MAST2 | 9423 | 18410 | -0.665 | 0.0111 | No |