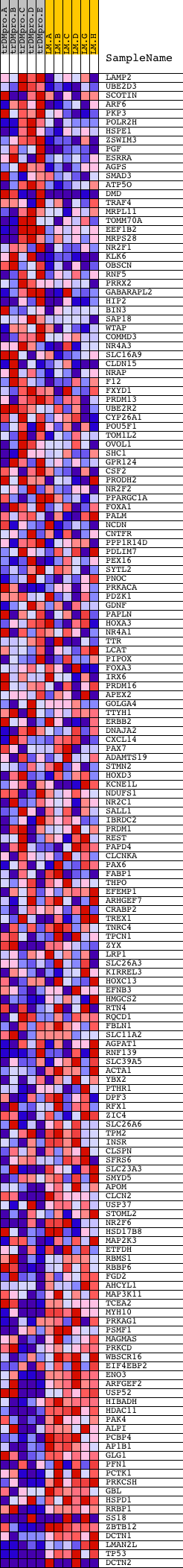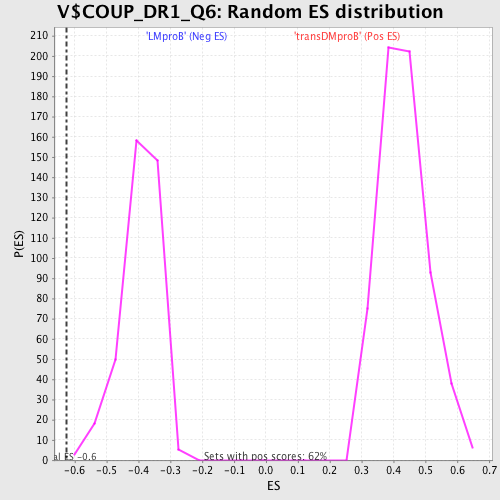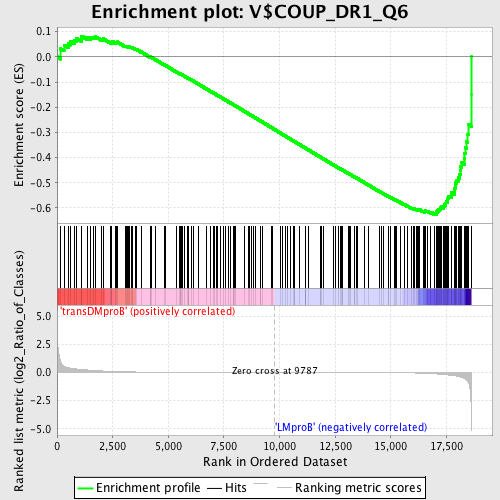
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$COUP_DR1_Q6 |
| Enrichment Score (ES) | -0.6257925 |
| Normalized Enrichment Score (NES) | -1.584212 |
| Nominal p-value | 0.002617801 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.568 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LAMP2 | 9267 2653 | 162 | 0.982 | 0.0316 | No | ||
| 2 | UBE2D3 | 7253 | 312 | 0.557 | 0.0464 | No | ||
| 3 | SCOTIN | 3136 19299 3102 | 508 | 0.408 | 0.0525 | No | ||
| 4 | ARF6 | 21252 | 612 | 0.367 | 0.0620 | No | ||
| 5 | PKP3 | 18016 | 786 | 0.319 | 0.0657 | No | ||
| 6 | POLR2H | 10888 | 865 | 0.302 | 0.0739 | No | ||
| 7 | HSPE1 | 9133 | 1100 | 0.259 | 0.0718 | No | ||
| 8 | ZSWIM3 | 14733 | 1104 | 0.258 | 0.0823 | No | ||
| 9 | PGF | 21020 | 1354 | 0.217 | 0.0777 | No | ||
| 10 | ESRRA | 23802 | 1519 | 0.193 | 0.0767 | No | ||
| 11 | AGPS | 14979 | 1619 | 0.178 | 0.0786 | No | ||
| 12 | SMAD3 | 19084 | 1706 | 0.168 | 0.0809 | No | ||
| 13 | ATP5O | 22539 | 1996 | 0.133 | 0.0707 | No | ||
| 14 | DMD | 24295 2647 | 2070 | 0.125 | 0.0719 | No | ||
| 15 | TRAF4 | 10217 5796 1400 | 2395 | 0.096 | 0.0583 | No | ||
| 16 | MRPL11 | 23774 | 2464 | 0.090 | 0.0583 | No | ||
| 17 | TOMM70A | 6608 | 2466 | 0.090 | 0.0619 | No | ||
| 18 | EEF1B2 | 4131 12063 | 2617 | 0.081 | 0.0571 | No | ||
| 19 | MRPS28 | 15377 | 2650 | 0.079 | 0.0586 | No | ||
| 20 | NR2F1 | 7015 21401 | 2698 | 0.075 | 0.0592 | No | ||
| 21 | KLK6 | 9231 9626 | 3057 | 0.055 | 0.0420 | No | ||
| 22 | OBSCN | 20434 | 3108 | 0.053 | 0.0415 | No | ||
| 23 | RNF5 | 63 | 3184 | 0.050 | 0.0395 | No | ||
| 24 | PRRX2 | 15047 | 3222 | 0.049 | 0.0395 | No | ||
| 25 | GABARAPL2 | 8211 13552 | 3243 | 0.048 | 0.0404 | No | ||
| 26 | HIP2 | 3626 3485 16838 | 3338 | 0.044 | 0.0371 | No | ||
| 27 | BIN3 | 21969 | 3402 | 0.042 | 0.0354 | No | ||
| 28 | SAP18 | 5406 | 3524 | 0.038 | 0.0304 | No | ||
| 29 | WTAP | 7215 | 3578 | 0.037 | 0.0291 | No | ||
| 30 | COMMD3 | 73 | 3772 | 0.032 | 0.0200 | No | ||
| 31 | NR4A3 | 9473 16212 5183 | 4183 | 0.024 | -0.0013 | No | ||
| 32 | SLC16A9 | 19993 | 4184 | 0.024 | -0.0003 | No | ||
| 33 | CLDN15 | 16667 3572 | 4258 | 0.023 | -0.0033 | No | ||
| 34 | NRAP | 23642 | 4435 | 0.021 | -0.0120 | No | ||
| 35 | F12 | 3260 21453 | 4808 | 0.017 | -0.0315 | No | ||
| 36 | FXYD1 | 17874 85 | 4888 | 0.016 | -0.0351 | No | ||
| 37 | PRDM13 | 15933 | 5358 | 0.013 | -0.0600 | No | ||
| 38 | UBE2R2 | 16240 | 5486 | 0.012 | -0.0664 | No | ||
| 39 | CYP26A1 | 23871 3695 | 5525 | 0.012 | -0.0679 | No | ||
| 40 | POU5F1 | 1547 23255 | 5531 | 0.012 | -0.0677 | No | ||
| 41 | TOM1L2 | 10115 | 5577 | 0.012 | -0.0697 | No | ||
| 42 | OVOL1 | 23983 | 5620 | 0.011 | -0.0715 | No | ||
| 43 | SHC1 | 9813 9812 5430 | 5728 | 0.011 | -0.0769 | No | ||
| 44 | GPR124 | 18643 918 | 5877 | 0.010 | -0.0845 | No | ||
| 45 | CSF2 | 20454 | 5902 | 0.010 | -0.0854 | No | ||
| 46 | PRODH2 | 1854 18300 | 5903 | 0.010 | -0.0850 | No | ||
| 47 | NR2F2 | 8619 409 | 6048 | 0.009 | -0.0924 | No | ||
| 48 | PPARGC1A | 16533 | 6148 | 0.009 | -0.0974 | No | ||
| 49 | FOXA1 | 21060 | 6372 | 0.008 | -0.1091 | No | ||
| 50 | PALM | 19960 3350 | 6698 | 0.007 | -0.1265 | No | ||
| 51 | NCDN | 15756 2487 6471 | 6897 | 0.006 | -0.1369 | No | ||
| 52 | CNTFR | 2515 15906 | 7043 | 0.006 | -0.1446 | No | ||
| 53 | PPP1R14D | 14471 | 7091 | 0.006 | -0.1469 | No | ||
| 54 | PDLIM7 | 7434 | 7185 | 0.006 | -0.1517 | No | ||
| 55 | PEX16 | 2681 | 7196 | 0.006 | -0.1520 | No | ||
| 56 | SYTL2 | 18190 1539 3730 | 7347 | 0.005 | -0.1599 | No | ||
| 57 | PNOC | 21783 | 7481 | 0.005 | -0.1669 | No | ||
| 58 | PRKACA | 18549 3844 | 7561 | 0.005 | -0.1710 | No | ||
| 59 | PDZK1 | 15490 | 7694 | 0.004 | -0.1780 | No | ||
| 60 | GDNF | 22514 2275 | 7813 | 0.004 | -0.1842 | No | ||
| 61 | PAPLN | 9301 | 7915 | 0.004 | -0.1896 | No | ||
| 62 | HOXA3 | 9108 1075 | 7917 | 0.004 | -0.1895 | No | ||
| 63 | NR4A1 | 9099 | 7973 | 0.003 | -0.1923 | No | ||
| 64 | TTR | 23615 | 8026 | 0.003 | -0.1950 | No | ||
| 65 | LCAT | 18762 | 8410 | 0.003 | -0.2156 | No | ||
| 66 | PIPOX | 20340 | 8618 | 0.002 | -0.2268 | No | ||
| 67 | FOXA3 | 17949 | 8664 | 0.002 | -0.2291 | No | ||
| 68 | IRX6 | 7225 | 8740 | 0.002 | -0.2331 | No | ||
| 69 | PRDM16 | 12893 | 8812 | 0.002 | -0.2369 | No | ||
| 70 | APEX2 | 8081 | 8894 | 0.002 | -0.2412 | No | ||
| 71 | GOLGA4 | 12022 | 9134 | 0.001 | -0.2541 | No | ||
| 72 | TTYH1 | 2496 7176 | 9143 | 0.001 | -0.2545 | No | ||
| 73 | ERBB2 | 8913 | 9244 | 0.001 | -0.2599 | No | ||
| 74 | DNAJA2 | 12133 18806 | 9629 | 0.000 | -0.2807 | No | ||
| 75 | CXCL14 | 21445 7165 | 9700 | 0.000 | -0.2844 | No | ||
| 76 | PAX7 | 15697 | 10052 | -0.001 | -0.3034 | No | ||
| 77 | ADAMTS19 | 23544 | 10133 | -0.001 | -0.3078 | No | ||
| 78 | STMN2 | 15638 | 10262 | -0.001 | -0.3147 | No | ||
| 79 | HOXD3 | 9115 | 10345 | -0.001 | -0.3191 | No | ||
| 80 | KCNE1L | 24044 | 10504 | -0.001 | -0.3276 | No | ||
| 81 | NDUFS1 | 13940 | 10638 | -0.002 | -0.3347 | No | ||
| 82 | NR2C1 | 10215 | 10666 | -0.002 | -0.3361 | No | ||
| 83 | SALL1 | 18796 12207 | 10678 | -0.002 | -0.3366 | No | ||
| 84 | IBRDC2 | 5747 | 10914 | -0.002 | -0.3493 | No | ||
| 85 | PRDM1 | 19775 3337 | 11162 | -0.003 | -0.3626 | No | ||
| 86 | REST | 16818 | 11167 | -0.003 | -0.3627 | No | ||
| 87 | PAPD4 | 8299 8300 4146 | 11168 | -0.003 | -0.3626 | No | ||
| 88 | CLCNKA | 15690 | 11299 | -0.003 | -0.3695 | No | ||
| 89 | PAX6 | 5223 | 11820 | -0.004 | -0.3975 | No | ||
| 90 | FABP1 | 17420 | 11822 | -0.004 | -0.3974 | No | ||
| 91 | THPO | 22636 | 11892 | -0.004 | -0.4010 | No | ||
| 92 | EFEMP1 | 20937 | 11969 | -0.004 | -0.4049 | No | ||
| 93 | ARHGEF7 | 3823 | 12410 | -0.005 | -0.4285 | No | ||
| 94 | CRABP2 | 15554 | 12493 | -0.006 | -0.4327 | No | ||
| 95 | TREX1 | 10219 3111 | 12666 | -0.006 | -0.4418 | No | ||
| 96 | TNRC4 | 15514 | 12743 | -0.006 | -0.4457 | No | ||
| 97 | TPCN1 | 16398 6383 10915 | 12766 | -0.006 | -0.4466 | No | ||
| 98 | ZYX | 10443 | 12825 | -0.007 | -0.4495 | No | ||
| 99 | LRP1 | 9284 | 12835 | -0.007 | -0.4497 | No | ||
| 100 | SLC26A3 | 21298 | 13082 | -0.007 | -0.4627 | No | ||
| 101 | KIRREL3 | 12571 | 13149 | -0.008 | -0.4660 | No | ||
| 102 | HOXC13 | 22344 | 13190 | -0.008 | -0.4678 | No | ||
| 103 | EFNB3 | 20391 | 13361 | -0.008 | -0.4767 | No | ||
| 104 | HMGCS2 | 15483 | 13367 | -0.008 | -0.4766 | No | ||
| 105 | RTN4 | 7569 1328 1343 1442 | 13468 | -0.009 | -0.4817 | No | ||
| 106 | RQCD1 | 12205 | 13504 | -0.009 | -0.4832 | No | ||
| 107 | FBLN1 | 4716 2208 | 13816 | -0.010 | -0.4997 | No | ||
| 108 | SLC11A2 | 22130 | 14008 | -0.011 | -0.5095 | No | ||
| 109 | AGPAT1 | 23275 | 14506 | -0.014 | -0.5359 | No | ||
| 110 | RNF139 | 7992 13291 | 14572 | -0.015 | -0.5388 | No | ||
| 111 | SLC39A5 | 3364 19596 | 14657 | -0.016 | -0.5427 | No | ||
| 112 | ACTA1 | 18715 | 14894 | -0.018 | -0.5548 | No | ||
| 113 | YBX2 | 20818 | 14912 | -0.018 | -0.5549 | No | ||
| 114 | PTHR1 | 5318 18985 | 14995 | -0.019 | -0.5586 | No | ||
| 115 | DPF3 | 7659 12853 | 15176 | -0.022 | -0.5674 | No | ||
| 116 | RFX1 | 18548 | 15226 | -0.023 | -0.5692 | No | ||
| 117 | ZIC4 | 5992 | 15237 | -0.023 | -0.5688 | No | ||
| 118 | SLC26A6 | 19304 | 15453 | -0.027 | -0.5793 | No | ||
| 119 | TPM2 | 2367 2437 | 15616 | -0.031 | -0.5868 | No | ||
| 120 | INSR | 18950 | 15741 | -0.035 | -0.5921 | No | ||
| 121 | CLSPN | 16079 11256 6530 | 15927 | -0.042 | -0.6003 | No | ||
| 122 | SFRS6 | 14751 | 15999 | -0.046 | -0.6023 | No | ||
| 123 | SLC23A3 | 10367 | 16057 | -0.048 | -0.6034 | No | ||
| 124 | SMYD5 | 17386 | 16144 | -0.053 | -0.6059 | No | ||
| 125 | APOM | 23007 | 16209 | -0.056 | -0.6071 | No | ||
| 126 | CLCN2 | 22637 | 16235 | -0.058 | -0.6061 | No | ||
| 127 | USP37 | 13923 | 16267 | -0.059 | -0.6053 | No | ||
| 128 | STOML2 | 15902 | 16451 | -0.072 | -0.6123 | No | ||
| 129 | NR2F6 | 4651 8876 18848 8912 | 16528 | -0.077 | -0.6132 | No | ||
| 130 | HSD17B8 | 9061 | 16547 | -0.079 | -0.6109 | No | ||
| 131 | MAP2K3 | 20856 | 16636 | -0.087 | -0.6121 | No | ||
| 132 | ETFDH | 15313 | 16787 | -0.102 | -0.6161 | No | ||
| 133 | RBMS1 | 14580 | 16958 | -0.120 | -0.6204 | Yes | ||
| 134 | RBBP6 | 5365 | 17059 | -0.133 | -0.6203 | Yes | ||
| 135 | FGD2 | 23309 | 17066 | -0.134 | -0.6151 | Yes | ||
| 136 | AHCYL1 | 6037 | 17099 | -0.139 | -0.6111 | Yes | ||
| 137 | MAP3K11 | 11163 | 17143 | -0.146 | -0.6075 | Yes | ||
| 138 | TCEA2 | 14699 | 17197 | -0.154 | -0.6041 | Yes | ||
| 139 | MYH10 | 8077 | 17236 | -0.160 | -0.5996 | Yes | ||
| 140 | PRKAG1 | 22135 | 17274 | -0.165 | -0.5948 | Yes | ||
| 141 | PSMF1 | 6009 | 17379 | -0.183 | -0.5929 | Yes | ||
| 142 | MAGMAS | 1677 22678 | 17411 | -0.189 | -0.5869 | Yes | ||
| 143 | PRKCD | 21897 | 17470 | -0.200 | -0.5818 | Yes | ||
| 144 | WBSCR16 | 16355 | 17480 | -0.202 | -0.5740 | Yes | ||
| 145 | EIF4EBP2 | 4662 | 17561 | -0.218 | -0.5694 | Yes | ||
| 146 | ENO3 | 8905 | 17566 | -0.219 | -0.5606 | Yes | ||
| 147 | ARFGEF2 | 14728 | 17608 | -0.228 | -0.5534 | Yes | ||
| 148 | USP52 | 3397 19841 | 17721 | -0.251 | -0.5492 | Yes | ||
| 149 | HIBADH | 17144 | 17732 | -0.253 | -0.5394 | Yes | ||
| 150 | HDAC11 | 17356 | 17846 | -0.286 | -0.5337 | Yes | ||
| 151 | PAK4 | 17909 | 17866 | -0.291 | -0.5228 | Yes | ||
| 152 | ALPI | 4892 | 17888 | -0.300 | -0.5116 | Yes | ||
| 153 | PCBP4 | 12235 | 17911 | -0.308 | -0.5002 | Yes | ||
| 154 | AP1B1 | 8599 | 17970 | -0.329 | -0.4898 | Yes | ||
| 155 | GLG1 | 5421 | 18052 | -0.362 | -0.4793 | Yes | ||
| 156 | PFN1 | 9555 | 18097 | -0.381 | -0.4661 | Yes | ||
| 157 | PCTK1 | 24369 | 18135 | -0.399 | -0.4517 | Yes | ||
| 158 | PRKCSH | 19535 | 18146 | -0.404 | -0.4357 | Yes | ||
| 159 | GBL | 23091 1612 | 18188 | -0.433 | -0.4201 | Yes | ||
| 160 | HSPD1 | 4078 9129 | 18315 | -0.533 | -0.4050 | Yes | ||
| 161 | RRBP1 | 8176 14411 | 18326 | -0.550 | -0.3830 | Yes | ||
| 162 | SS18 | 6506 6505 2030 | 18356 | -0.579 | -0.3607 | Yes | ||
| 163 | ZBTB12 | 23268 | 18395 | -0.637 | -0.3366 | Yes | ||
| 164 | DCTN1 | 1100 17392 | 18464 | -0.790 | -0.3079 | Yes | ||
| 165 | LMAN2L | 5670 10070 5669 | 18507 | -0.990 | -0.2695 | Yes | ||
| 166 | TP53 | 20822 | 18610 | -3.019 | -0.1510 | Yes | ||
| 167 | DCTN2 | 7635 12812 | 18612 | -3.684 | 0.0002 | Yes |