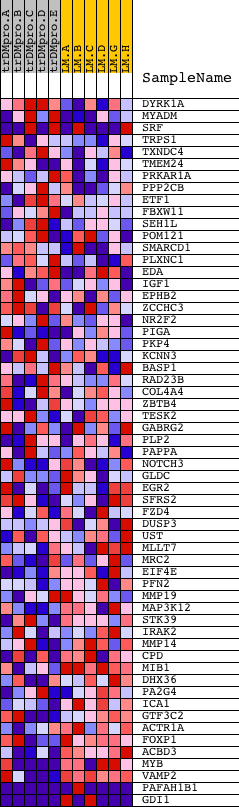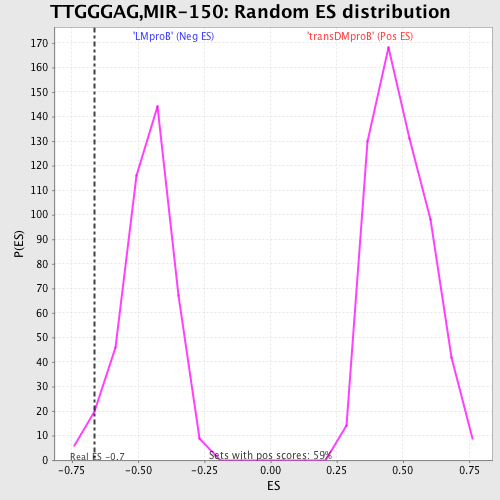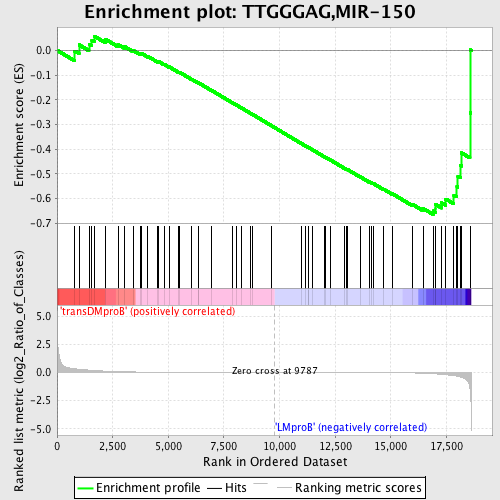
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | TTGGGAG,MIR-150 |
| Enrichment Score (ES) | -0.66427577 |
| Normalized Enrichment Score (NES) | -1.4267576 |
| Nominal p-value | 0.039215688 |
| FDR q-value | 0.5174223 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DYRK1A | 4649 | 794 | 0.317 | -0.0027 | No | ||
| 2 | MYADM | 11946 | 988 | 0.280 | 0.0224 | No | ||
| 3 | SRF | 22961 1597 | 1434 | 0.207 | 0.0246 | No | ||
| 4 | TRPS1 | 8195 | 1563 | 0.187 | 0.0414 | No | ||
| 5 | TXNDC4 | 15886 | 1668 | 0.172 | 0.0576 | No | ||
| 6 | TMEM24 | 19150 | 2185 | 0.115 | 0.0443 | No | ||
| 7 | PRKAR1A | 20615 | 2739 | 0.073 | 0.0237 | No | ||
| 8 | PPP2CB | 18636 | 3031 | 0.057 | 0.0152 | No | ||
| 9 | ETF1 | 23467 | 3435 | 0.041 | -0.0013 | No | ||
| 10 | FBXW11 | 20926 | 3738 | 0.033 | -0.0134 | No | ||
| 11 | SEH1L | 13001 | 3776 | 0.032 | -0.0113 | No | ||
| 12 | POM121 | 16343 | 4077 | 0.026 | -0.0242 | No | ||
| 13 | SMARCD1 | 2241 22360 2250 | 4522 | 0.020 | -0.0456 | No | ||
| 14 | PLXNC1 | 7056 | 4553 | 0.019 | -0.0448 | No | ||
| 15 | EDA | 24284 | 4822 | 0.017 | -0.0571 | No | ||
| 16 | IGF1 | 3352 9156 3409 | 5037 | 0.015 | -0.0668 | No | ||
| 17 | EPHB2 | 4675 2440 8910 | 5459 | 0.012 | -0.0879 | No | ||
| 18 | ZCCHC3 | 12602 7504 12603 | 5489 | 0.012 | -0.0880 | No | ||
| 19 | NR2F2 | 8619 409 | 6048 | 0.009 | -0.1169 | No | ||
| 20 | PIGA | 2592 24210 | 6338 | 0.008 | -0.1314 | No | ||
| 21 | PKP4 | 2822 15008 | 6346 | 0.008 | -0.1308 | No | ||
| 22 | KCNN3 | 15538 1913 | 6918 | 0.006 | -0.1607 | No | ||
| 23 | BASP1 | 7668 | 7877 | 0.004 | -0.2119 | No | ||
| 24 | RAD23B | 5349 | 8043 | 0.003 | -0.2203 | No | ||
| 25 | COL4A4 | 8767 | 8275 | 0.003 | -0.2324 | No | ||
| 26 | ZBTB4 | 13267 | 8684 | 0.002 | -0.2542 | No | ||
| 27 | TESK2 | 10504 | 8796 | 0.002 | -0.2599 | No | ||
| 28 | GABRG2 | 1259 4748 8996 1331 | 9637 | 0.000 | -0.3051 | No | ||
| 29 | PLP2 | 9591 | 10991 | -0.002 | -0.3777 | No | ||
| 30 | PAPPA | 16190 | 11171 | -0.003 | -0.3871 | No | ||
| 31 | NOTCH3 | 23031 | 11303 | -0.003 | -0.3937 | No | ||
| 32 | GLDC | 23701 | 11319 | -0.003 | -0.3942 | No | ||
| 33 | EGR2 | 8886 | 11481 | -0.003 | -0.4025 | No | ||
| 34 | SFRS2 | 9807 20136 | 12005 | -0.004 | -0.4301 | No | ||
| 35 | FZD4 | 18193 | 12070 | -0.005 | -0.4329 | No | ||
| 36 | DUSP3 | 1358 13022 | 12288 | -0.005 | -0.4440 | No | ||
| 37 | UST | 6865 | 12922 | -0.007 | -0.4772 | No | ||
| 38 | MLLT7 | 24278 | 13026 | -0.007 | -0.4819 | No | ||
| 39 | MRC2 | 1386 5117 | 13072 | -0.007 | -0.4833 | No | ||
| 40 | EIF4E | 15403 1827 8890 | 13623 | -0.009 | -0.5118 | No | ||
| 41 | PFN2 | 15337 1818 | 14032 | -0.011 | -0.5323 | No | ||
| 42 | MMP19 | 7191 | 14116 | -0.012 | -0.5353 | No | ||
| 43 | MAP3K12 | 6454 11164 | 14211 | -0.012 | -0.5388 | No | ||
| 44 | STK39 | 14570 | 14678 | -0.016 | -0.5619 | No | ||
| 45 | IRAK2 | 17324 | 15073 | -0.021 | -0.5806 | No | ||
| 46 | MMP14 | 22020 | 15971 | -0.044 | -0.6233 | No | ||
| 47 | CPD | 20344 4556 | 16454 | -0.072 | -0.6401 | No | ||
| 48 | MIB1 | 5908 | 16903 | -0.114 | -0.6499 | Yes | ||
| 49 | DHX36 | 15328 | 17010 | -0.126 | -0.6396 | Yes | ||
| 50 | PA2G4 | 5265 | 17012 | -0.127 | -0.6236 | Yes | ||
| 51 | ICA1 | 17217 | 17283 | -0.166 | -0.6171 | Yes | ||
| 52 | GTF3C2 | 7749 | 17468 | -0.199 | -0.6018 | Yes | ||
| 53 | ACTR1A | 23653 | 17834 | -0.281 | -0.5859 | Yes | ||
| 54 | FOXP1 | 4242 | 17971 | -0.329 | -0.5515 | Yes | ||
| 55 | ACBD3 | 14025 | 18015 | -0.347 | -0.5099 | Yes | ||
| 56 | MYB | 3344 3355 19806 | 18127 | -0.395 | -0.4658 | Yes | ||
| 57 | VAMP2 | 20826 | 18175 | -0.423 | -0.4147 | Yes | ||
| 58 | PAFAH1B1 | 1340 5220 9524 | 18559 | -1.441 | -0.2530 | Yes | ||
| 59 | GDI1 | 9012 4770 24297 4769 | 18581 | -2.022 | 0.0019 | Yes |