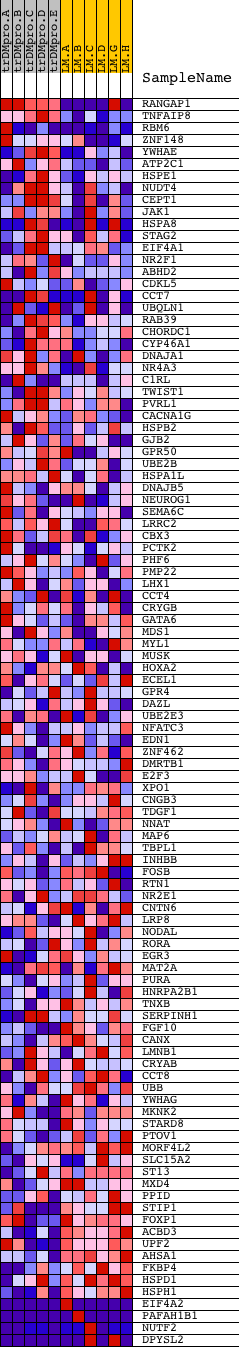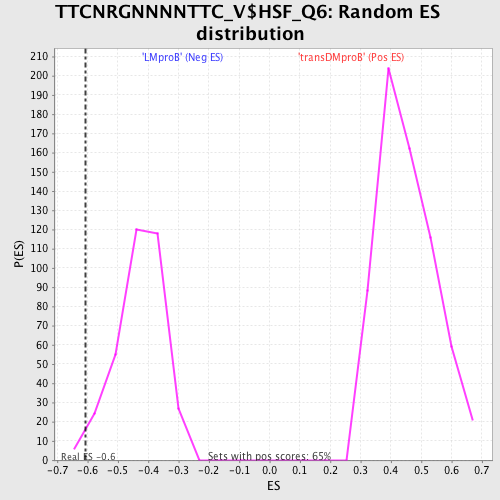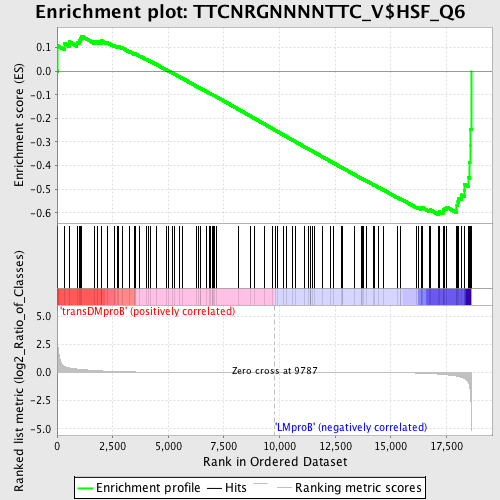
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | TTCNRGNNNNTTC_V$HSF_Q6 |
| Enrichment Score (ES) | -0.6072593 |
| Normalized Enrichment Score (NES) | -1.4203959 |
| Nominal p-value | 0.02 |
| FDR q-value | 0.51926124 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RANGAP1 | 2180 22195 | 37 | 2.246 | 0.1071 | No | ||
| 2 | TNFAIP8 | 8391 | 342 | 0.532 | 0.1165 | No | ||
| 3 | RBM6 | 9707 3095 3063 2995 | 543 | 0.394 | 0.1249 | No | ||
| 4 | ZNF148 | 5949 | 893 | 0.297 | 0.1204 | No | ||
| 5 | YWHAE | 20776 | 986 | 0.281 | 0.1291 | No | ||
| 6 | ATP2C1 | 10660 6178 | 1043 | 0.269 | 0.1392 | No | ||
| 7 | HSPE1 | 9133 | 1100 | 0.259 | 0.1487 | No | ||
| 8 | NUDT4 | 19639 | 1678 | 0.171 | 0.1259 | No | ||
| 9 | CEPT1 | 13624 | 1815 | 0.155 | 0.1261 | No | ||
| 10 | JAK1 | 15827 | 1982 | 0.134 | 0.1236 | No | ||
| 11 | HSPA8 | 9124 3072 4871 | 2007 | 0.132 | 0.1287 | No | ||
| 12 | STAG2 | 5521 | 2243 | 0.109 | 0.1214 | No | ||
| 13 | EIF4A1 | 8889 23719 | 2569 | 0.084 | 0.1079 | No | ||
| 14 | NR2F1 | 7015 21401 | 2698 | 0.075 | 0.1046 | No | ||
| 15 | ABHD2 | 18208 12043 | 2779 | 0.070 | 0.1037 | No | ||
| 16 | CDKL5 | 24019 | 2917 | 0.063 | 0.0993 | No | ||
| 17 | CCT7 | 17385 | 3271 | 0.047 | 0.0825 | No | ||
| 18 | UBQLN1 | 3237 3209 3248 3291 | 3458 | 0.041 | 0.0745 | No | ||
| 19 | RAB39 | 19113 | 3518 | 0.039 | 0.0731 | No | ||
| 20 | CHORDC1 | 12430 | 3715 | 0.034 | 0.0642 | No | ||
| 21 | CYP46A1 | 21158 | 4024 | 0.027 | 0.0489 | No | ||
| 22 | DNAJA1 | 4878 16242 2345 | 4121 | 0.025 | 0.0449 | No | ||
| 23 | NR4A3 | 9473 16212 5183 | 4183 | 0.024 | 0.0428 | No | ||
| 24 | C1RL | 10559 17288 | 4482 | 0.020 | 0.0277 | No | ||
| 25 | TWIST1 | 21291 | 4910 | 0.016 | 0.0054 | No | ||
| 26 | PVRL1 | 3006 19482 12211 7192 | 5015 | 0.015 | 0.0005 | No | ||
| 27 | CACNA1G | 1237 940 20292 | 5184 | 0.014 | -0.0079 | No | ||
| 28 | HSPB2 | 19121 | 5288 | 0.013 | -0.0129 | No | ||
| 29 | GJB2 | 9018 21806 | 5490 | 0.012 | -0.0231 | No | ||
| 30 | GPR50 | 24318 | 5633 | 0.011 | -0.0303 | No | ||
| 31 | UBE2B | 10245 | 6252 | 0.008 | -0.0632 | No | ||
| 32 | HSPA1L | 23267 | 6351 | 0.008 | -0.0681 | No | ||
| 33 | DNAJB5 | 16236 | 6455 | 0.008 | -0.0733 | No | ||
| 34 | NEUROG1 | 21446 | 6459 | 0.008 | -0.0731 | No | ||
| 35 | SEMA6C | 1797 5424 | 6693 | 0.007 | -0.0854 | No | ||
| 36 | LRRC2 | 13189 | 6870 | 0.006 | -0.0945 | No | ||
| 37 | CBX3 | 8705 4484 | 6890 | 0.006 | -0.0953 | No | ||
| 38 | PCTK2 | 19907 | 6971 | 0.006 | -0.0993 | No | ||
| 39 | PHF6 | 2648 24340 | 6975 | 0.006 | -0.0992 | No | ||
| 40 | PMP22 | 9594 1312 | 7046 | 0.006 | -0.1027 | No | ||
| 41 | LHX1 | 4995 | 7064 | 0.006 | -0.1033 | No | ||
| 42 | CCT4 | 8710 | 7149 | 0.006 | -0.1076 | No | ||
| 43 | CRYGB | 8795 | 8131 | 0.003 | -0.1604 | No | ||
| 44 | GATA6 | 96 | 8682 | 0.002 | -0.1900 | No | ||
| 45 | MDS1 | 9375 | 8858 | 0.002 | -0.1994 | No | ||
| 46 | MYL1 | 4015 | 9319 | 0.001 | -0.2242 | No | ||
| 47 | MUSK | 5199 | 9693 | 0.000 | -0.2443 | No | ||
| 48 | HOXA2 | 17151 | 9807 | -0.000 | -0.2504 | No | ||
| 49 | ECEL1 | 13893 | 9907 | -0.000 | -0.2557 | No | ||
| 50 | GPR4 | 18364 | 10191 | -0.001 | -0.2710 | No | ||
| 51 | DAZL | 22944 | 10312 | -0.001 | -0.2774 | No | ||
| 52 | UBE2E3 | 5816 5815 | 10576 | -0.001 | -0.2916 | No | ||
| 53 | NFATC3 | 5169 | 10711 | -0.002 | -0.2987 | No | ||
| 54 | EDN1 | 21658 | 11116 | -0.003 | -0.3204 | No | ||
| 55 | ZNF462 | 6321 | 11320 | -0.003 | -0.3312 | No | ||
| 56 | DMRTB1 | 15813 | 11376 | -0.003 | -0.3341 | No | ||
| 57 | E2F3 | 21500 8874 | 11384 | -0.003 | -0.3343 | No | ||
| 58 | XPO1 | 4172 | 11397 | -0.003 | -0.3348 | No | ||
| 59 | CNGB3 | 16267 | 11474 | -0.003 | -0.3387 | No | ||
| 60 | TDGF1 | 5704 10108 | 11574 | -0.003 | -0.3439 | No | ||
| 61 | NNAT | 2764 5180 | 11950 | -0.004 | -0.3640 | No | ||
| 62 | MAP6 | 9419 | 12308 | -0.005 | -0.3830 | No | ||
| 63 | TBPL1 | 19805 | 12407 | -0.005 | -0.3880 | No | ||
| 64 | INHBB | 13858 | 12799 | -0.006 | -0.4088 | No | ||
| 65 | FOSB | 17945 | 12838 | -0.007 | -0.4106 | No | ||
| 66 | RTN1 | 4177 2099 | 12844 | -0.007 | -0.4105 | No | ||
| 67 | NR2E1 | 19780 | 13354 | -0.008 | -0.4376 | No | ||
| 68 | CNTN6 | 17346 | 13663 | -0.010 | -0.4538 | No | ||
| 69 | LRP8 | 16149 2425 | 13687 | -0.010 | -0.4545 | No | ||
| 70 | NODAL | 333 20010 | 13721 | -0.010 | -0.4558 | No | ||
| 71 | RORA | 3019 5388 | 13769 | -0.010 | -0.4579 | No | ||
| 72 | EGR3 | 4656 | 13887 | -0.011 | -0.4637 | No | ||
| 73 | MAT2A | 10553 10554 6099 | 14240 | -0.013 | -0.4821 | No | ||
| 74 | PURA | 9670 | 14263 | -0.013 | -0.4827 | No | ||
| 75 | HNRPA2B1 | 1187 7001 7000 1040 | 14459 | -0.014 | -0.4925 | No | ||
| 76 | TNXB | 1557 8174 878 8175 | 14663 | -0.016 | -0.5027 | No | ||
| 77 | SERPINH1 | 8703 | 15283 | -0.024 | -0.5350 | No | ||
| 78 | FGF10 | 4719 8962 | 15428 | -0.027 | -0.5415 | No | ||
| 79 | CANX | 4474 | 15443 | -0.027 | -0.5409 | No | ||
| 80 | LMNB1 | 23549 1943 | 16150 | -0.053 | -0.5765 | No | ||
| 81 | CRYAB | 19457 | 16264 | -0.059 | -0.5797 | No | ||
| 82 | CCT8 | 22551 | 16358 | -0.065 | -0.5815 | No | ||
| 83 | UBB | 10240 | 16381 | -0.067 | -0.5795 | No | ||
| 84 | YWHAG | 16339 | 16425 | -0.070 | -0.5784 | No | ||
| 85 | MKNK2 | 3299 9392 | 16718 | -0.094 | -0.5895 | No | ||
| 86 | STARD8 | 10682 | 16762 | -0.099 | -0.5871 | No | ||
| 87 | PTOV1 | 13522 | 17137 | -0.144 | -0.6002 | Yes | ||
| 88 | MORF4L2 | 12118 | 17189 | -0.152 | -0.5956 | Yes | ||
| 89 | SLC15A2 | 22600 | 17347 | -0.178 | -0.5955 | Yes | ||
| 90 | ST13 | 12865 | 17363 | -0.180 | -0.5875 | Yes | ||
| 91 | MXD4 | 9346 | 17431 | -0.192 | -0.5818 | Yes | ||
| 92 | PPID | 12576 | 17514 | -0.209 | -0.5761 | Yes | ||
| 93 | STIP1 | 23798 | 17969 | -0.329 | -0.5846 | Yes | ||
| 94 | FOXP1 | 4242 | 17971 | -0.329 | -0.5687 | Yes | ||
| 95 | ACBD3 | 14025 | 18015 | -0.347 | -0.5541 | Yes | ||
| 96 | UPF2 | 11591 | 18058 | -0.365 | -0.5387 | Yes | ||
| 97 | AHSA1 | 21192 | 18183 | -0.428 | -0.5246 | Yes | ||
| 98 | FKBP4 | 4726 | 18310 | -0.528 | -0.5057 | Yes | ||
| 99 | HSPD1 | 4078 9129 | 18315 | -0.533 | -0.4801 | Yes | ||
| 100 | HSPH1 | 16282 | 18472 | -0.807 | -0.4493 | Yes | ||
| 101 | EIF4A2 | 4660 1679 1645 | 18550 | -1.408 | -0.3850 | Yes | ||
| 102 | PAFAH1B1 | 1340 5220 9524 | 18559 | -1.441 | -0.3154 | Yes | ||
| 103 | NUTF2 | 12632 7529 | 18561 | -1.450 | -0.2450 | Yes | ||
| 104 | DPYSL2 | 3122 4561 8787 | 18616 | -5.104 | -0.0000 | Yes |