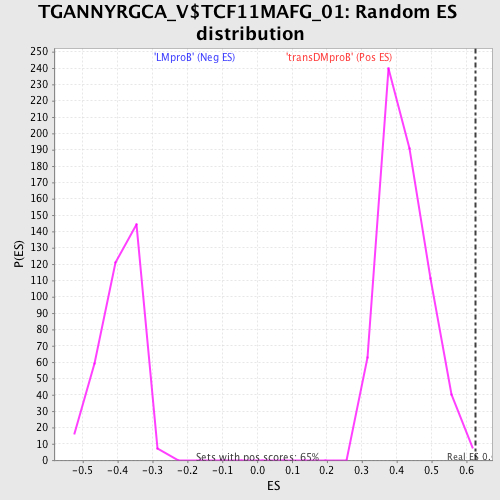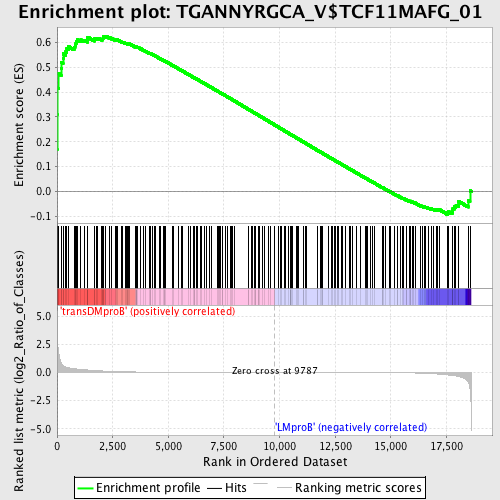
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | TGANNYRGCA_V$TCF11MAFG_01 |
| Enrichment Score (ES) | 0.62529415 |
| Normalized Enrichment Score (NES) | 1.4785975 |
| Nominal p-value | 0.0030627872 |
| FDR q-value | 0.4889811 |
| FWER p-Value | 0.992 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NDUFC1 | 7287 12339 | 0 | 5.436 | 0.1700 | Yes | ||
| 2 | RAN | 5356 9691 | 6 | 4.449 | 0.3089 | Yes | ||
| 3 | TPM3 | 12233 7209 7208 1790 | 12 | 3.461 | 0.4168 | Yes | ||
| 4 | MMP9 | 14732 | 56 | 1.927 | 0.4747 | Yes | ||
| 5 | BCL6 | 22624 | 181 | 0.872 | 0.4953 | Yes | ||
| 6 | RAG1 | 9689 | 192 | 0.809 | 0.5200 | Yes | ||
| 7 | PSMD8 | 7166 | 267 | 0.625 | 0.5355 | Yes | ||
| 8 | DTX2 | 16674 3488 | 278 | 0.610 | 0.5541 | Yes | ||
| 9 | NEDD9 | 3206 21483 | 392 | 0.481 | 0.5630 | Yes | ||
| 10 | POP1 | 2214 7482 | 437 | 0.450 | 0.5747 | Yes | ||
| 11 | SERTAD4 | 13715 | 513 | 0.406 | 0.5833 | Yes | ||
| 12 | TXNL1 | 7002 | 764 | 0.324 | 0.5799 | Yes | ||
| 13 | BACH1 | 8647 | 806 | 0.315 | 0.5875 | Yes | ||
| 14 | IPO11 | 21353 | 843 | 0.307 | 0.5952 | Yes | ||
| 15 | PSMB1 | 23118 | 857 | 0.304 | 0.6039 | Yes | ||
| 16 | RAB6A | 18173 3905 | 900 | 0.296 | 0.6109 | Yes | ||
| 17 | SNX10 | 17445 | 1069 | 0.265 | 0.6101 | Yes | ||
| 18 | GSTP1 | 9051 4812 4811 | 1220 | 0.235 | 0.6093 | Yes | ||
| 19 | TOB1 | 20703 | 1368 | 0.216 | 0.6081 | Yes | ||
| 20 | BLMH | 20769 | 1369 | 0.216 | 0.6148 | Yes | ||
| 21 | MCART1 | 10492 | 1372 | 0.216 | 0.6215 | Yes | ||
| 22 | RBBP7 | 10887 6366 10886 | 1667 | 0.172 | 0.6109 | Yes | ||
| 23 | TBL1X | 5628 | 1695 | 0.169 | 0.6147 | Yes | ||
| 24 | RHOG | 17727 | 1773 | 0.160 | 0.6156 | Yes | ||
| 25 | EEF2 | 8881 4654 8882 | 1829 | 0.154 | 0.6174 | Yes | ||
| 26 | CIRBP | 8742 | 1979 | 0.135 | 0.6135 | Yes | ||
| 27 | RPL23 | 7237 12267 20267 | 2054 | 0.127 | 0.6135 | Yes | ||
| 28 | SQSTM1 | 9517 | 2059 | 0.126 | 0.6172 | Yes | ||
| 29 | DMD | 24295 2647 | 2070 | 0.125 | 0.6206 | Yes | ||
| 30 | GADD45G | 21637 | 2077 | 0.124 | 0.6241 | Yes | ||
| 31 | GEMIN7 | 17944 | 2186 | 0.115 | 0.6218 | Yes | ||
| 32 | ARRDC3 | 4191 | 2189 | 0.114 | 0.6253 | Yes | ||
| 33 | GADD45A | 17129 | 2367 | 0.098 | 0.6187 | No | ||
| 34 | TOMM70A | 6608 | 2466 | 0.090 | 0.6162 | No | ||
| 35 | MYO1C | 20777 | 2631 | 0.080 | 0.6098 | No | ||
| 36 | SPTLC2 | 5505 5506 | 2676 | 0.076 | 0.6098 | No | ||
| 37 | ABCB6 | 13162 | 2692 | 0.075 | 0.6114 | No | ||
| 38 | PTGES2 | 15038 | 2905 | 0.063 | 0.6018 | No | ||
| 39 | AP1G1 | 4392 | 2951 | 0.061 | 0.6013 | No | ||
| 40 | PSMC5 | 1348 20624 | 3086 | 0.054 | 0.5957 | No | ||
| 41 | ACO1 | 16244 | 3112 | 0.053 | 0.5960 | No | ||
| 42 | HINT1 | 20881 | 3177 | 0.050 | 0.5941 | No | ||
| 43 | UCHL1 | 16834 | 3182 | 0.050 | 0.5954 | No | ||
| 44 | DUSP13 | 21904 | 3189 | 0.050 | 0.5967 | No | ||
| 45 | SLC35B1 | 1631 23226 | 3258 | 0.047 | 0.5945 | No | ||
| 46 | IQGAP1 | 6619 | 3500 | 0.039 | 0.5826 | No | ||
| 47 | VASP | 5847 | 3549 | 0.038 | 0.5812 | No | ||
| 48 | FBXO44 | 15675 | 3556 | 0.038 | 0.5820 | No | ||
| 49 | NKIRAS2 | 12979 | 3574 | 0.037 | 0.5823 | No | ||
| 50 | SOX5 | 9850 1044 5480 16937 | 3612 | 0.036 | 0.5814 | No | ||
| 51 | ROM1 | 23751 | 3766 | 0.033 | 0.5741 | No | ||
| 52 | KCNA6 | 664 16990 665 4938 | 3875 | 0.030 | 0.5692 | No | ||
| 53 | GRM7 | 17338 | 3990 | 0.027 | 0.5639 | No | ||
| 54 | ALDOB | 10494 | 4162 | 0.024 | 0.5553 | No | ||
| 55 | NR4A3 | 9473 16212 5183 | 4183 | 0.024 | 0.5550 | No | ||
| 56 | ANGPTL4 | 1504 7178 | 4201 | 0.024 | 0.5548 | No | ||
| 57 | ABCF3 | 6578 | 4284 | 0.022 | 0.5511 | No | ||
| 58 | PPARG | 1151 1144 17319 | 4359 | 0.021 | 0.5477 | No | ||
| 59 | PTPRN | 13914 | 4418 | 0.021 | 0.5452 | No | ||
| 60 | SAMD7 | 7998 13298 | 4598 | 0.019 | 0.5361 | No | ||
| 61 | MCAM | 3109 19479 | 4665 | 0.018 | 0.5331 | No | ||
| 62 | SUMO2 | 1293 9319 | 4792 | 0.017 | 0.5268 | No | ||
| 63 | HRSP12 | 9123 9122 4870 9638 | 4810 | 0.017 | 0.5264 | No | ||
| 64 | RASAL1 | 16719 | 4847 | 0.016 | 0.5249 | No | ||
| 65 | KLHL11 | 20229 | 4868 | 0.016 | 0.5243 | No | ||
| 66 | LRP1B | 8249 | 5206 | 0.014 | 0.5065 | No | ||
| 67 | TBXAS1 | 17480 1105 | 5210 | 0.014 | 0.5067 | No | ||
| 68 | NANOS1 | 6858 | 5453 | 0.012 | 0.4940 | No | ||
| 69 | MAPK12 | 2182 2215 22164 | 5589 | 0.011 | 0.4870 | No | ||
| 70 | ARF4 | 22072 | 5629 | 0.011 | 0.4853 | No | ||
| 71 | KCNH3 | 9207 | 5924 | 0.010 | 0.4696 | No | ||
| 72 | CDC45L | 22642 1752 | 5980 | 0.010 | 0.4669 | No | ||
| 73 | TLR8 | 9308 | 6124 | 0.009 | 0.4594 | No | ||
| 74 | MYOC | 9440 14079 | 6158 | 0.009 | 0.4579 | No | ||
| 75 | CST7 | 14805 | 6196 | 0.009 | 0.4562 | No | ||
| 76 | SEC23A | 5420 | 6248 | 0.008 | 0.4537 | No | ||
| 77 | KCND1 | 4940 | 6318 | 0.008 | 0.4502 | No | ||
| 78 | SNCB | 21456 | 6451 | 0.008 | 0.4433 | No | ||
| 79 | ANGPT1 | 8588 2260 22303 | 6479 | 0.008 | 0.4420 | No | ||
| 80 | MAPK10 | 11169 | 6611 | 0.007 | 0.4351 | No | ||
| 81 | FGF1 | 1994 23447 | 6735 | 0.007 | 0.4287 | No | ||
| 82 | SLC22A18 | 18000 | 6839 | 0.007 | 0.4233 | No | ||
| 83 | FLNC | 4302 8510 | 6933 | 0.006 | 0.4184 | No | ||
| 84 | DHDDS | 2536 15724 | 7202 | 0.005 | 0.4041 | No | ||
| 85 | CDON | 12195 | 7233 | 0.005 | 0.4026 | No | ||
| 86 | NAP1L5 | 17134 | 7290 | 0.005 | 0.3997 | No | ||
| 87 | PFKFB1 | 9552 | 7348 | 0.005 | 0.3968 | No | ||
| 88 | CDK9 | 14619 | 7415 | 0.005 | 0.3934 | No | ||
| 89 | FCHSD1 | 6661 11435 | 7441 | 0.005 | 0.3922 | No | ||
| 90 | ABCC6 | 17823 | 7562 | 0.005 | 0.3858 | No | ||
| 91 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 7640 | 0.004 | 0.3817 | No | ||
| 92 | CPNE6 | 22010 | 7804 | 0.004 | 0.3730 | No | ||
| 93 | MYL3 | 19291 | 7837 | 0.004 | 0.3714 | No | ||
| 94 | SLC7A8 | 3611 21833 | 7871 | 0.004 | 0.3697 | No | ||
| 95 | BNIP3 | 17581 | 7984 | 0.003 | 0.3637 | No | ||
| 96 | KCNAB1 | 1791 15577 | 8583 | 0.002 | 0.3314 | No | ||
| 97 | RNF13 | 6271 | 8621 | 0.002 | 0.3294 | No | ||
| 98 | TCF1 | 16416 | 8726 | 0.002 | 0.3238 | No | ||
| 99 | RYR1 | 17903 17902 9770 | 8797 | 0.002 | 0.3201 | No | ||
| 100 | FOXP2 | 4312 17523 8523 | 8879 | 0.002 | 0.3157 | No | ||
| 101 | TLL1 | 10188 | 8909 | 0.002 | 0.3142 | No | ||
| 102 | TINAG | 3141 19060 | 8913 | 0.002 | 0.3141 | No | ||
| 103 | HDLBP | 4017 13873 | 9049 | 0.001 | 0.3068 | No | ||
| 104 | CD2AP | 22975 | 9053 | 0.001 | 0.3067 | No | ||
| 105 | OSBP | 4278 | 9100 | 0.001 | 0.3043 | No | ||
| 106 | NAP1L2 | 24087 | 9237 | 0.001 | 0.2969 | No | ||
| 107 | CACNB1 | 1443 1304 | 9321 | 0.001 | 0.2924 | No | ||
| 108 | DUSP2 | 14865 | 9504 | 0.001 | 0.2826 | No | ||
| 109 | NUDT11 | 24387 | 9572 | 0.000 | 0.2789 | No | ||
| 110 | RHO | 17314 | 9750 | 0.000 | 0.2693 | No | ||
| 111 | SYT2 | 14122 | 9786 | 0.000 | 0.2674 | No | ||
| 112 | SPG3A | 7878 13145 | 9970 | -0.000 | 0.2575 | No | ||
| 113 | CAMKK1 | 20793 | 10058 | -0.001 | 0.2528 | No | ||
| 114 | DOCK4 | 10705 | 10063 | -0.001 | 0.2526 | No | ||
| 115 | XLKD1 | 17675 | 10205 | -0.001 | 0.2450 | No | ||
| 116 | STMN2 | 15638 | 10262 | -0.001 | 0.2420 | No | ||
| 117 | LTA | 23003 | 10388 | -0.001 | 0.2352 | No | ||
| 118 | HOXB8 | 9111 | 10475 | -0.001 | 0.2306 | No | ||
| 119 | CRH | 8784 | 10491 | -0.001 | 0.2298 | No | ||
| 120 | NRG1 | 3885 | 10499 | -0.001 | 0.2295 | No | ||
| 121 | ESR1 | 20097 4685 | 10530 | -0.001 | 0.2279 | No | ||
| 122 | HSPB3 | 21340 | 10561 | -0.001 | 0.2263 | No | ||
| 123 | PADI4 | 2424 9547 | 10768 | -0.002 | 0.2152 | No | ||
| 124 | GNG3 | 23752 | 10785 | -0.002 | 0.2144 | No | ||
| 125 | KDELR2 | 16634 12428 | 10834 | -0.002 | 0.2118 | No | ||
| 126 | KCTD4 | 21956 | 11065 | -0.002 | 0.1994 | No | ||
| 127 | FTSJ3 | 20187 | 11094 | -0.002 | 0.1980 | No | ||
| 128 | PRDM1 | 19775 3337 | 11162 | -0.003 | 0.1944 | No | ||
| 129 | P2RXL1 | 5218 9521 | 11214 | -0.003 | 0.1917 | No | ||
| 130 | SERPINB5 | 9861 | 11685 | -0.004 | 0.1663 | No | ||
| 131 | SMOX | 10455 2940 2737 2952 | 11691 | -0.004 | 0.1662 | No | ||
| 132 | CPZ | 16557 | 11816 | -0.004 | 0.1596 | No | ||
| 133 | SIX6 | 21247 | 11864 | -0.004 | 0.1572 | No | ||
| 134 | FGF9 | 8966 | 11912 | -0.004 | 0.1547 | No | ||
| 135 | CLTC | 14610 | 11919 | -0.004 | 0.1545 | No | ||
| 136 | APBA2BP | 14385 | 12208 | -0.005 | 0.1391 | No | ||
| 137 | SSR1 | 4211 | 12340 | -0.005 | 0.1321 | No | ||
| 138 | SMPX | 24220 | 12364 | -0.005 | 0.1310 | No | ||
| 139 | DTNA | 8870 4647 23611 1991 | 12455 | -0.006 | 0.1263 | No | ||
| 140 | TFEC | 17213 | 12513 | -0.006 | 0.1234 | No | ||
| 141 | RORC | 9732 | 12597 | -0.006 | 0.1191 | No | ||
| 142 | SDPR | 14252 | 12651 | -0.006 | 0.1164 | No | ||
| 143 | PSMA1 | 1627 17669 | 12796 | -0.006 | 0.1088 | No | ||
| 144 | LRP1 | 9284 | 12835 | -0.007 | 0.1069 | No | ||
| 145 | WDFY3 | 7789 3607 16462 | 12983 | -0.007 | 0.0992 | No | ||
| 146 | ADAM23 | 10699 4057 | 13161 | -0.008 | 0.0898 | No | ||
| 147 | HNRPR | 13196 7909 2500 | 13188 | -0.008 | 0.0886 | No | ||
| 148 | FGF11 | 20383 | 13286 | -0.008 | 0.0836 | No | ||
| 149 | GAB2 | 1821 18184 2025 | 13442 | -0.009 | 0.0755 | No | ||
| 150 | CLCN5 | 24204 | 13658 | -0.010 | 0.0641 | No | ||
| 151 | TYRO3 | 5811 | 13862 | -0.011 | 0.0534 | No | ||
| 152 | DRP2 | 24254 | 13896 | -0.011 | 0.0519 | No | ||
| 153 | TBP | 671 1554 | 13937 | -0.011 | 0.0501 | No | ||
| 154 | UBE2D1 | 19738 | 14099 | -0.012 | 0.0417 | No | ||
| 155 | RARG | 22113 | 14154 | -0.012 | 0.0392 | No | ||
| 156 | IL6 | 16895 | 14243 | -0.013 | 0.0348 | No | ||
| 157 | SLC8A3 | 2065 4293 | 14619 | -0.015 | 0.0149 | No | ||
| 158 | SPATS2 | 13044 | 14621 | -0.015 | 0.0153 | No | ||
| 159 | FBXO2 | 15993 | 14648 | -0.016 | 0.0144 | No | ||
| 160 | ASPSCR1 | 12751 1300 | 14769 | -0.017 | 0.0084 | No | ||
| 161 | CRYGN | 16587 | 14956 | -0.019 | -0.0011 | No | ||
| 162 | PPP2CA | 20890 | 14993 | -0.019 | -0.0024 | No | ||
| 163 | OMG | 5209 | 15158 | -0.022 | -0.0107 | No | ||
| 164 | MAMDC1 | 21054 6798 | 15299 | -0.024 | -0.0175 | No | ||
| 165 | SNTB2 | 3887 18476 | 15317 | -0.025 | -0.0177 | No | ||
| 166 | ABCA7 | 3308 19952 | 15421 | -0.027 | -0.0224 | No | ||
| 167 | MTMR4 | 20712 | 15520 | -0.029 | -0.0268 | No | ||
| 168 | BAZ2A | 4371 | 15560 | -0.030 | -0.0280 | No | ||
| 169 | UFD1L | 22825 | 15711 | -0.035 | -0.0351 | No | ||
| 170 | FBLN2 | 994 8956 | 15713 | -0.035 | -0.0341 | No | ||
| 171 | CFL2 | 2155 21069 | 15840 | -0.039 | -0.0397 | No | ||
| 172 | MAPK3 | 6458 11170 | 15861 | -0.039 | -0.0395 | No | ||
| 173 | FMNL1 | 12191 | 15866 | -0.040 | -0.0385 | No | ||
| 174 | PSMD11 | 12772 7600 | 15961 | -0.044 | -0.0423 | No | ||
| 175 | SEC24D | 15429 | 16023 | -0.047 | -0.0441 | No | ||
| 176 | BTK | 24061 | 16092 | -0.050 | -0.0462 | No | ||
| 177 | ENO1 | 8903 | 16334 | -0.064 | -0.0573 | No | ||
| 178 | YWHAG | 16339 | 16425 | -0.070 | -0.0600 | No | ||
| 179 | PIM1 | 23308 | 16509 | -0.076 | -0.0621 | No | ||
| 180 | BSCL2 | 23940 | 16572 | -0.081 | -0.0629 | No | ||
| 181 | NR6A1 | 14596 | 16714 | -0.094 | -0.0676 | No | ||
| 182 | ANP32E | 15500 | 16819 | -0.104 | -0.0700 | No | ||
| 183 | BRD2 | 4735 8984 7628 | 16901 | -0.114 | -0.0709 | No | ||
| 184 | ATP2A3 | 20795 1314 | 17031 | -0.129 | -0.0738 | No | ||
| 185 | SLC16A6 | 4186 | 17079 | -0.136 | -0.0721 | No | ||
| 186 | PSMB2 | 2324 16078 | 17171 | -0.149 | -0.0724 | No | ||
| 187 | GPD1 | 22361 | 17548 | -0.217 | -0.0860 | No | ||
| 188 | ATP1B1 | 4420 | 17583 | -0.223 | -0.0809 | No | ||
| 189 | CMAS | 17245 | 17756 | -0.259 | -0.0822 | No | ||
| 190 | MYST1 | 18059 | 17781 | -0.267 | -0.0751 | No | ||
| 191 | VCP | 11254 | 17790 | -0.268 | -0.0672 | No | ||
| 192 | EIF5 | 5736 | 17859 | -0.289 | -0.0618 | No | ||
| 193 | RABGGTB | 15134 | 17928 | -0.314 | -0.0557 | No | ||
| 194 | DNAJC7 | 20227 | 18043 | -0.357 | -0.0507 | No | ||
| 195 | GABARAPL1 | 17268 | 18048 | -0.360 | -0.0397 | No | ||
| 196 | PSMD7 | 18752 3825 | 18474 | -0.810 | -0.0374 | No | ||
| 197 | PAFAH1B1 | 1340 5220 9524 | 18559 | -1.441 | 0.0031 | No |