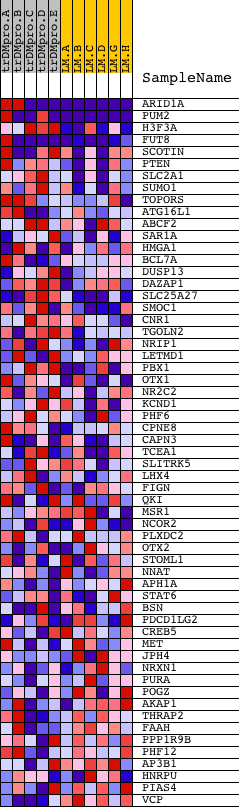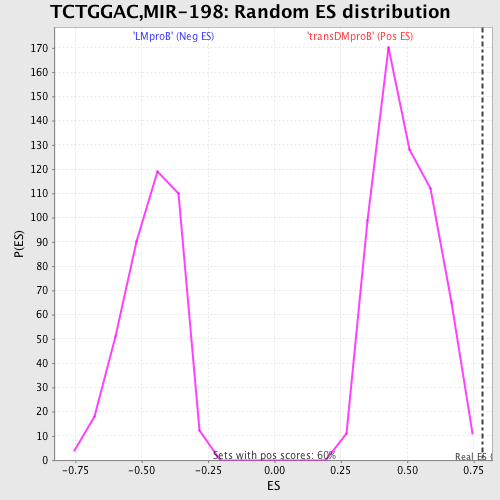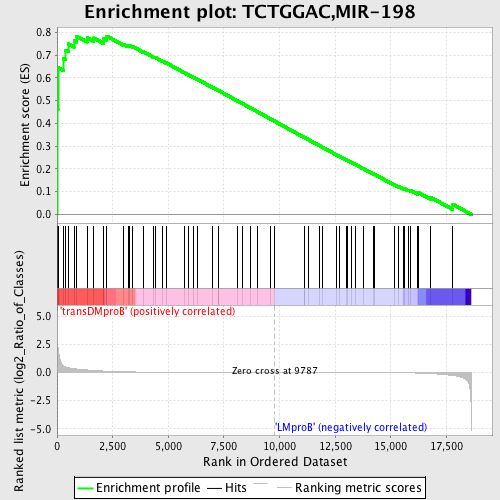
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | TCTGGAC,MIR-198 |
| Enrichment Score (ES) | 0.7836079 |
| Normalized Enrichment Score (NES) | 1.5956503 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15294707 |
| FWER p-Value | 0.405 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 1 | 5.436 | 0.4644 | Yes | ||
| 2 | PUM2 | 2071 8161 | 41 | 2.138 | 0.6450 | Yes | ||
| 3 | H3F3A | 4838 | 277 | 0.613 | 0.6847 | Yes | ||
| 4 | FUT8 | 2163 21233 | 386 | 0.491 | 0.7209 | Yes | ||
| 5 | SCOTIN | 3136 19299 3102 | 508 | 0.408 | 0.7492 | Yes | ||
| 6 | PTEN | 5305 | 762 | 0.324 | 0.7633 | Yes | ||
| 7 | SLC2A1 | 16107 | 870 | 0.301 | 0.7832 | Yes | ||
| 8 | SUMO1 | 5826 3943 10247 | 1345 | 0.219 | 0.7764 | Yes | ||
| 9 | TOPORS | 15920 | 1641 | 0.174 | 0.7754 | Yes | ||
| 10 | ATG16L1 | 14197 | 2095 | 0.122 | 0.7614 | Yes | ||
| 11 | ABCF2 | 11338 6580 6579 | 2102 | 0.121 | 0.7714 | Yes | ||
| 12 | SAR1A | 20008 | 2218 | 0.113 | 0.7749 | Yes | ||
| 13 | HMGA1 | 23323 | 2232 | 0.111 | 0.7836 | Yes | ||
| 14 | BCL7A | 8063 | 2993 | 0.059 | 0.7477 | No | ||
| 15 | DUSP13 | 21904 | 3189 | 0.050 | 0.7414 | No | ||
| 16 | DAZAP1 | 19945 | 3241 | 0.048 | 0.7428 | No | ||
| 17 | SLC25A27 | 7880 13149 | 3403 | 0.042 | 0.7377 | No | ||
| 18 | SMOC1 | 21221 | 3883 | 0.030 | 0.7145 | No | ||
| 19 | CNR1 | 4537 2362 | 4347 | 0.021 | 0.6914 | No | ||
| 20 | TGOLN2 | 10230 | 4409 | 0.021 | 0.6899 | No | ||
| 21 | NRIP1 | 11216 | 4748 | 0.017 | 0.6731 | No | ||
| 22 | LETMD1 | 22357 | 4903 | 0.016 | 0.6662 | No | ||
| 23 | PBX1 | 5224 | 5707 | 0.011 | 0.6239 | No | ||
| 24 | OTX1 | 20517 | 5923 | 0.010 | 0.6131 | No | ||
| 25 | NR2C2 | 10216 | 6131 | 0.009 | 0.6027 | No | ||
| 26 | KCND1 | 4940 | 6318 | 0.008 | 0.5934 | No | ||
| 27 | PHF6 | 2648 24340 | 6975 | 0.006 | 0.5586 | No | ||
| 28 | CPNE8 | 22156 | 7255 | 0.005 | 0.5440 | No | ||
| 29 | CAPN3 | 8686 2868 | 7269 | 0.005 | 0.5437 | No | ||
| 30 | TCEA1 | 10043 | 8114 | 0.003 | 0.4985 | No | ||
| 31 | SLITRK5 | 21939 | 8328 | 0.003 | 0.4873 | No | ||
| 32 | LHX4 | 13800 | 8709 | 0.002 | 0.4670 | No | ||
| 33 | FIGN | 14575 | 8985 | 0.002 | 0.4523 | No | ||
| 34 | QKI | 23128 5340 | 9609 | 0.000 | 0.4188 | No | ||
| 35 | MSR1 | 5417 | 9781 | 0.000 | 0.4096 | No | ||
| 36 | NCOR2 | 9835 16367 | 11101 | -0.003 | 0.3387 | No | ||
| 37 | PLXDC2 | 15114 | 11137 | -0.003 | 0.3370 | No | ||
| 38 | OTX2 | 9519 | 11293 | -0.003 | 0.3289 | No | ||
| 39 | STOML1 | 3062 19424 | 11792 | -0.004 | 0.3024 | No | ||
| 40 | NNAT | 2764 5180 | 11950 | -0.004 | 0.2943 | No | ||
| 41 | APH1A | 15501 5947 10378 | 12546 | -0.006 | 0.2628 | No | ||
| 42 | STAT6 | 19854 9909 | 12671 | -0.006 | 0.2566 | No | ||
| 43 | BSN | 18997 | 13022 | -0.007 | 0.2384 | No | ||
| 44 | PDCD1LG2 | 23892 | 13058 | -0.007 | 0.2371 | No | ||
| 45 | CREB5 | 10551 | 13237 | -0.008 | 0.2282 | No | ||
| 46 | MET | 17520 | 13399 | -0.008 | 0.2202 | No | ||
| 47 | JPH4 | 21826 | 13765 | -0.010 | 0.2014 | No | ||
| 48 | NRXN1 | 1581 1575 22875 | 14207 | -0.012 | 0.1787 | No | ||
| 49 | PURA | 9670 | 14263 | -0.013 | 0.1768 | No | ||
| 50 | POGZ | 6031 1911 | 15149 | -0.022 | 0.1310 | No | ||
| 51 | AKAP1 | 4364 1326 1286 20299 | 15352 | -0.025 | 0.1223 | No | ||
| 52 | THRAP2 | 8009 | 15551 | -0.029 | 0.1141 | No | ||
| 53 | FAAH | 4707 8942 | 15611 | -0.031 | 0.1136 | No | ||
| 54 | PPP1R9B | 20698 | 15789 | -0.037 | 0.1072 | No | ||
| 55 | PHF12 | 11192 6483 | 15889 | -0.041 | 0.1054 | No | ||
| 56 | AP3B1 | 21583 | 16219 | -0.057 | 0.0925 | No | ||
| 57 | HNRPU | 6959 | 16221 | -0.057 | 0.0973 | No | ||
| 58 | PIAS4 | 19681 | 16803 | -0.103 | 0.0748 | No | ||
| 59 | VCP | 11254 | 17790 | -0.268 | 0.0445 | No |