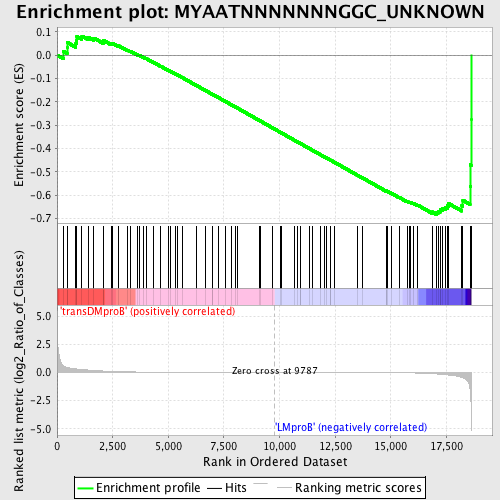
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | MYAATNNNNNNNGGC_UNKNOWN |
| Enrichment Score (ES) | -0.68189687 |
| Normalized Enrichment Score (NES) | -1.56211 |
| Nominal p-value | 0.007575758 |
| FDR q-value | 0.7284388 |
| FWER p-Value | 0.685 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SQLE | 22468 2229 | 282 | 0.592 | 0.0165 | No | ||
| 2 | NFKB2 | 23810 | 451 | 0.442 | 0.0312 | No | ||
| 3 | ACAA2 | 1956 1981 23515 | 458 | 0.439 | 0.0544 | No | ||
| 4 | FLI1 | 4729 | 841 | 0.307 | 0.0502 | No | ||
| 5 | CDC20 | 8421 8422 4227 | 875 | 0.299 | 0.0645 | No | ||
| 6 | BCAS3 | 5314 | 887 | 0.298 | 0.0798 | No | ||
| 7 | PAPOLG | 20512 | 1113 | 0.256 | 0.0814 | No | ||
| 8 | TLE4 | 23720 3697 | 1401 | 0.211 | 0.0772 | No | ||
| 9 | TMSB10 | 5322 | 1655 | 0.174 | 0.0729 | No | ||
| 10 | DMD | 24295 2647 | 2070 | 0.125 | 0.0573 | No | ||
| 11 | AMD1 | 8583 | 2090 | 0.122 | 0.0628 | No | ||
| 12 | FKBP2 | 8972 | 2437 | 0.093 | 0.0491 | No | ||
| 13 | RKHD1 | 10693 | 2509 | 0.088 | 0.0500 | No | ||
| 14 | RABAC1 | 1407 9007 | 2757 | 0.072 | 0.0405 | No | ||
| 15 | GPX1 | 19310 | 3178 | 0.050 | 0.0205 | No | ||
| 16 | BCL2L1 | 4440 2930 8652 | 3290 | 0.046 | 0.0170 | No | ||
| 17 | SOX5 | 9850 1044 5480 16937 | 3612 | 0.036 | 0.0016 | No | ||
| 18 | SYT4 | 5566 | 3704 | 0.034 | -0.0015 | No | ||
| 19 | KCNA6 | 664 16990 665 4938 | 3875 | 0.030 | -0.0090 | No | ||
| 20 | SWAP70 | 18126 5553 9948 | 3996 | 0.027 | -0.0141 | No | ||
| 21 | HOXD4 | 4866 | 4329 | 0.022 | -0.0308 | No | ||
| 22 | EIF2C1 | 10672 | 4651 | 0.018 | -0.0471 | No | ||
| 23 | PVRL1 | 3006 19482 12211 7192 | 5015 | 0.015 | -0.0659 | No | ||
| 24 | RNF38 | 7862 2328 13115 | 5108 | 0.014 | -0.0701 | No | ||
| 25 | VSX1 | 2766 14398 | 5339 | 0.013 | -0.0818 | No | ||
| 26 | TLE1 | 5765 10185 15859 | 5397 | 0.013 | -0.0842 | No | ||
| 27 | LUC7L2 | 5313 5312 | 5613 | 0.011 | -0.0952 | No | ||
| 28 | RXRG | 14063 | 6283 | 0.008 | -0.1308 | No | ||
| 29 | LBX1 | 98 23659 | 6672 | 0.007 | -0.1514 | No | ||
| 30 | PCTK2 | 19907 | 6971 | 0.006 | -0.1672 | No | ||
| 31 | RPS6KA5 | 2076 21005 | 6999 | 0.006 | -0.1683 | No | ||
| 32 | JAG1 | 14415 | 7246 | 0.005 | -0.1813 | No | ||
| 33 | PRKACA | 18549 3844 | 7561 | 0.005 | -0.1980 | No | ||
| 34 | LAMA1 | 23168 | 7849 | 0.004 | -0.2132 | No | ||
| 35 | PTK7 | 22959 | 8032 | 0.003 | -0.2229 | No | ||
| 36 | NRXN3 | 2153 5196 | 8109 | 0.003 | -0.2268 | No | ||
| 37 | OSBP | 4278 | 9100 | 0.001 | -0.2801 | No | ||
| 38 | KCNIP4 | 3638 16536 | 9138 | 0.001 | -0.2821 | No | ||
| 39 | CXCL14 | 21445 7165 | 9700 | 0.000 | -0.3123 | No | ||
| 40 | ASCL2 | 5079 | 10044 | -0.001 | -0.3308 | No | ||
| 41 | DOCK4 | 10705 | 10063 | -0.001 | -0.3317 | No | ||
| 42 | SALL1 | 18796 12207 | 10678 | -0.002 | -0.3648 | No | ||
| 43 | USP15 | 4758 3377 | 10782 | -0.002 | -0.3702 | No | ||
| 44 | DAB1 | 4594 8834 | 10937 | -0.002 | -0.3784 | No | ||
| 45 | BBOX1 | 5010 9290 | 10960 | -0.002 | -0.3795 | No | ||
| 46 | PAQR6 | 15551 12756 7591 | 11334 | -0.003 | -0.3995 | No | ||
| 47 | TITF1 | 10180 21062 | 11473 | -0.003 | -0.4067 | No | ||
| 48 | PAX6 | 5223 | 11820 | -0.004 | -0.4252 | No | ||
| 49 | GABRA6 | 20491 | 12023 | -0.004 | -0.4358 | No | ||
| 50 | BDNF | 14926 2797 | 12108 | -0.005 | -0.4401 | No | ||
| 51 | SOST | 20210 | 12292 | -0.005 | -0.4497 | No | ||
| 52 | POU3F3 | 14261 | 12297 | -0.005 | -0.4497 | No | ||
| 53 | ARHGEF12 | 19156 | 12482 | -0.006 | -0.4593 | No | ||
| 54 | NSD1 | 2134 5197 | 13515 | -0.009 | -0.5145 | No | ||
| 55 | FGF3 | 17997 | 13743 | -0.010 | -0.5262 | No | ||
| 56 | NFYA | 5172 | 14783 | -0.017 | -0.5813 | No | ||
| 57 | TNPO2 | 5608 | 14788 | -0.017 | -0.5807 | No | ||
| 58 | SLC26A9 | 11560 980 4067 | 14859 | -0.018 | -0.5835 | No | ||
| 59 | WASL | 17203 | 15021 | -0.020 | -0.5911 | No | ||
| 60 | PCYT1B | 24289 | 15389 | -0.026 | -0.6095 | No | ||
| 61 | ETV5 | 22630 | 15738 | -0.035 | -0.6264 | No | ||
| 62 | LIN28 | 15723 | 15852 | -0.039 | -0.6304 | No | ||
| 63 | PANK2 | 14838 | 15878 | -0.040 | -0.6296 | No | ||
| 64 | SFRS6 | 14751 | 15999 | -0.046 | -0.6336 | No | ||
| 65 | PPP2R5D | 962 1523 22957 10139 | 16199 | -0.056 | -0.6414 | No | ||
| 66 | CENPF | 13723 | 16855 | -0.107 | -0.6709 | No | ||
| 67 | RBBP6 | 5365 | 17059 | -0.133 | -0.6747 | Yes | ||
| 68 | PLA2G6 | 22209 | 17123 | -0.143 | -0.6705 | Yes | ||
| 69 | TYK2 | 12058 19215 | 17212 | -0.157 | -0.6668 | Yes | ||
| 70 | FBXL20 | 13010 | 17233 | -0.159 | -0.6594 | Yes | ||
| 71 | XBP1 | 10318 | 17334 | -0.177 | -0.6553 | Yes | ||
| 72 | GTF3C2 | 7749 | 17468 | -0.199 | -0.6518 | Yes | ||
| 73 | TULP3 | 16984 | 17546 | -0.216 | -0.6444 | Yes | ||
| 74 | NDST2 | 21906 | 17596 | -0.225 | -0.6350 | Yes | ||
| 75 | DLX1 | 14988 | 18193 | -0.436 | -0.6437 | Yes | ||
| 76 | FKBP5 | 23051 | 18239 | -0.466 | -0.6212 | Yes | ||
| 77 | NUTF2 | 12632 7529 | 18561 | -1.450 | -0.5607 | Yes | ||
| 78 | ANP32A | 8592 4387 19415 | 18574 | -1.725 | -0.4689 | Yes | ||
| 79 | DCTN2 | 7635 12812 | 18612 | -3.684 | -0.2734 | Yes | ||
| 80 | DPYSL2 | 3122 4561 8787 | 18616 | -5.104 | -0.0000 | Yes |

