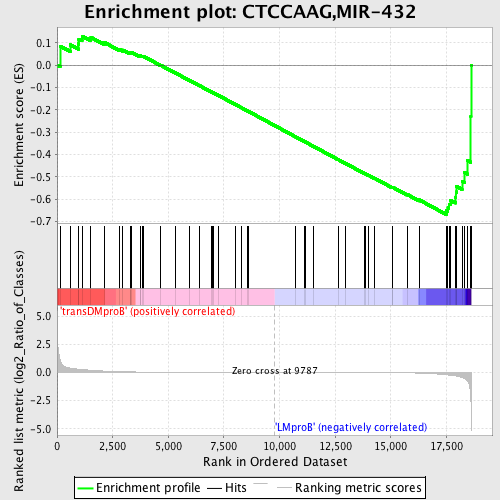
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | CTCCAAG,MIR-432 |
| Enrichment Score (ES) | -0.66825604 |
| Normalized Enrichment Score (NES) | -1.4459952 |
| Nominal p-value | 0.025943397 |
| FDR q-value | 0.5284708 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EHD3 | 23159 | 135 | 1.135 | 0.0854 | No | ||
| 2 | EML4 | 8122 | 589 | 0.374 | 0.0916 | No | ||
| 3 | ARL5B | 15115 | 954 | 0.287 | 0.0954 | No | ||
| 4 | AP3S1 | 8604 | 976 | 0.283 | 0.1173 | No | ||
| 5 | AMFR | 6209 | 1122 | 0.254 | 0.1303 | No | ||
| 6 | ESRRA | 23802 | 1519 | 0.193 | 0.1247 | No | ||
| 7 | CSE1L | 2906 | 2118 | 0.120 | 0.1023 | No | ||
| 8 | PLAGL2 | 7055 | 2811 | 0.068 | 0.0706 | No | ||
| 9 | RASGRP4 | 6119 | 2949 | 0.061 | 0.0682 | No | ||
| 10 | DAB2IP | 12808 2692 | 3276 | 0.047 | 0.0544 | No | ||
| 11 | ACE | 1419 4313 | 3300 | 0.046 | 0.0569 | No | ||
| 12 | SFRS1 | 8492 | 3328 | 0.045 | 0.0591 | No | ||
| 13 | FURIN | 9538 | 3733 | 0.033 | 0.0401 | No | ||
| 14 | ABCC3 | 20293 | 3740 | 0.033 | 0.0425 | No | ||
| 15 | GRIA2 | 4802 | 3815 | 0.032 | 0.0411 | No | ||
| 16 | CELSR2 | 15194 12000 | 3869 | 0.030 | 0.0407 | No | ||
| 17 | EIF2C1 | 10672 | 4651 | 0.018 | 0.0001 | No | ||
| 18 | SPARC | 20444 | 5304 | 0.013 | -0.0339 | No | ||
| 19 | CRB3 | 23176 | 5962 | 0.010 | -0.0685 | No | ||
| 20 | FN1 | 4091 4094 971 13929 | 6406 | 0.008 | -0.0918 | No | ||
| 21 | LUZP1 | 6533 11257 2503 | 6958 | 0.006 | -0.1209 | No | ||
| 22 | PRPF4B | 3208 5295 | 7003 | 0.006 | -0.1228 | No | ||
| 23 | PMP22 | 9594 1312 | 7046 | 0.006 | -0.1246 | No | ||
| 24 | CAPN3 | 8686 2868 | 7269 | 0.005 | -0.1361 | No | ||
| 25 | STK35 | 14851 | 8020 | 0.003 | -0.1763 | No | ||
| 26 | PITPNC1 | 7759 | 8278 | 0.003 | -0.1899 | No | ||
| 27 | HSPA14 | 14695 | 8538 | 0.002 | -0.2036 | No | ||
| 28 | SMCR8 | 53 | 8580 | 0.002 | -0.2056 | No | ||
| 29 | HOMER2 | 11184 | 10693 | -0.002 | -0.3193 | No | ||
| 30 | NR5A2 | 4105 13817 | 11131 | -0.003 | -0.3426 | No | ||
| 31 | TRPC3 | 15355 | 11149 | -0.003 | -0.3433 | No | ||
| 32 | CPEB2 | 6077 | 11540 | -0.003 | -0.3641 | No | ||
| 33 | SDPR | 14252 | 12651 | -0.006 | -0.4234 | No | ||
| 34 | HIF1AN | 23840 | 12949 | -0.007 | -0.4388 | No | ||
| 35 | ABCA9 | 1452 20166 | 13804 | -0.010 | -0.4839 | No | ||
| 36 | TRIM9 | 2118 8076 13577 8234 | 13841 | -0.010 | -0.4850 | No | ||
| 37 | CA3 | 4476 8690 | 13982 | -0.011 | -0.4917 | No | ||
| 38 | PURA | 9670 | 14263 | -0.013 | -0.5057 | No | ||
| 39 | BRPF1 | 17332 | 15053 | -0.020 | -0.5466 | No | ||
| 40 | HMGA2 | 4857 9098 3341 3444 | 15088 | -0.021 | -0.5467 | No | ||
| 41 | PDE4A | 3037 19542 3083 3023 | 15728 | -0.035 | -0.5783 | No | ||
| 42 | DCTN3 | 15908 | 16279 | -0.060 | -0.6030 | No | ||
| 43 | POLE3 | 7200 | 17492 | -0.205 | -0.6515 | Yes | ||
| 44 | HOXA5 | 17149 | 17569 | -0.220 | -0.6376 | Yes | ||
| 45 | CPT1B | 2187 22159 | 17614 | -0.229 | -0.6213 | Yes | ||
| 46 | BACE1 | 6214 | 17690 | -0.244 | -0.6054 | Yes | ||
| 47 | POLDIP2 | 20754 | 17905 | -0.307 | -0.5919 | Yes | ||
| 48 | FNTB | 21235 | 17929 | -0.315 | -0.5674 | Yes | ||
| 49 | TLN1 | 15899 | 17946 | -0.323 | -0.5419 | Yes | ||
| 50 | RNF44 | 8364 | 18204 | -0.441 | -0.5197 | Yes | ||
| 51 | CHKB | 4518 | 18321 | -0.547 | -0.4813 | Yes | ||
| 52 | PDE1B | 9542 | 18452 | -0.747 | -0.4273 | Yes | ||
| 53 | SLMAP | 8198 13521 2609 | 18598 | -2.531 | -0.2284 | Yes | ||
| 54 | NFAT5 | 3921 7037 12036 | 18604 | -2.808 | 0.0006 | Yes |

