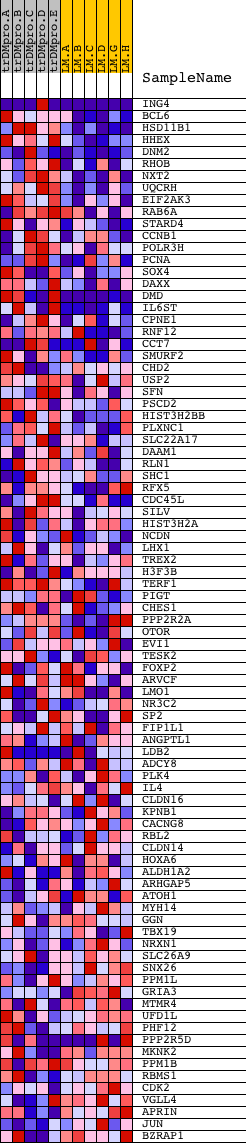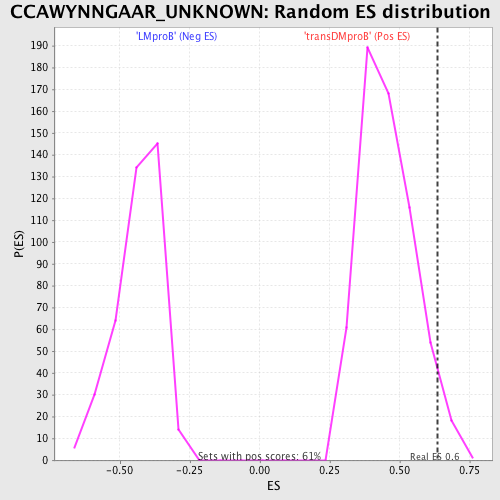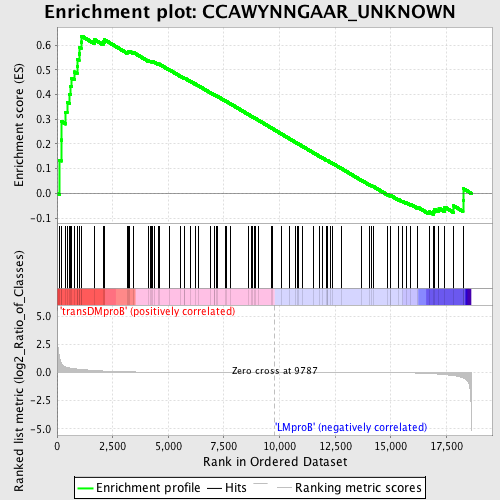
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | CCAWYNNGAAR_UNKNOWN |
| Enrichment Score (ES) | 0.6360157 |
| Normalized Enrichment Score (NES) | 1.3952928 |
| Nominal p-value | 0.03789127 |
| FDR q-value | 0.62484777 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ING4 | 6602 1155 1102 11371 | 104 | 1.439 | 0.1350 | Yes | ||
| 2 | BCL6 | 22624 | 181 | 0.872 | 0.2160 | Yes | ||
| 3 | HSD11B1 | 9125 4019 | 205 | 0.771 | 0.2901 | Yes | ||
| 4 | HHEX | 23872 | 384 | 0.493 | 0.3287 | Yes | ||
| 5 | DNM2 | 4635 3103 | 449 | 0.443 | 0.3685 | Yes | ||
| 6 | RHOB | 4411 | 555 | 0.390 | 0.4010 | Yes | ||
| 7 | NXT2 | 10685 | 607 | 0.368 | 0.4341 | Yes | ||
| 8 | UQCRH | 12378 | 654 | 0.356 | 0.4664 | Yes | ||
| 9 | EIF2AK3 | 17421 | 777 | 0.320 | 0.4911 | Yes | ||
| 10 | RAB6A | 18173 3905 | 900 | 0.296 | 0.5134 | Yes | ||
| 11 | STARD4 | 5011 | 923 | 0.292 | 0.5407 | Yes | ||
| 12 | CCNB1 | 11201 21362 | 987 | 0.280 | 0.5647 | Yes | ||
| 13 | POLR3H | 13460 8135 | 1020 | 0.274 | 0.5897 | Yes | ||
| 14 | PCNA | 9535 | 1079 | 0.263 | 0.6123 | Yes | ||
| 15 | SOX4 | 5479 | 1106 | 0.257 | 0.6360 | Yes | ||
| 16 | DAXX | 23290 | 1676 | 0.171 | 0.6221 | No | ||
| 17 | DMD | 24295 2647 | 2070 | 0.125 | 0.6130 | No | ||
| 18 | IL6ST | 4920 | 2133 | 0.119 | 0.6213 | No | ||
| 19 | CPNE1 | 11186 | 3146 | 0.052 | 0.5717 | No | ||
| 20 | RNF12 | 5384 | 3191 | 0.050 | 0.5742 | No | ||
| 21 | CCT7 | 17385 | 3271 | 0.047 | 0.5745 | No | ||
| 22 | SMURF2 | 185 | 3428 | 0.042 | 0.5701 | No | ||
| 23 | CHD2 | 10847 | 4086 | 0.026 | 0.5372 | No | ||
| 24 | USP2 | 19480 3052 3043 | 4193 | 0.024 | 0.5338 | No | ||
| 25 | SFN | 7060 12062 | 4246 | 0.023 | 0.5333 | No | ||
| 26 | PSCD2 | 17826 | 4283 | 0.022 | 0.5335 | No | ||
| 27 | HIST3H2BB | 11796 | 4371 | 0.021 | 0.5309 | No | ||
| 28 | PLXNC1 | 7056 | 4553 | 0.019 | 0.5230 | No | ||
| 29 | SLC22A17 | 21830 3096 | 4581 | 0.019 | 0.5234 | No | ||
| 30 | DAAM1 | 5536 | 5063 | 0.015 | 0.4989 | No | ||
| 31 | RLN1 | 9726 | 5565 | 0.012 | 0.4730 | No | ||
| 32 | SHC1 | 9813 9812 5430 | 5728 | 0.011 | 0.4653 | No | ||
| 33 | RFX5 | 7018 | 5730 | 0.011 | 0.4663 | No | ||
| 34 | CDC45L | 22642 1752 | 5980 | 0.010 | 0.4538 | No | ||
| 35 | SILV | 16130 | 6197 | 0.009 | 0.4430 | No | ||
| 36 | HIST3H2A | 6647 | 6334 | 0.008 | 0.4364 | No | ||
| 37 | NCDN | 15756 2487 6471 | 6897 | 0.006 | 0.4067 | No | ||
| 38 | LHX1 | 4995 | 7064 | 0.006 | 0.3983 | No | ||
| 39 | TREX2 | 24137 | 7076 | 0.006 | 0.3983 | No | ||
| 40 | H3F3B | 4839 | 7182 | 0.006 | 0.3932 | No | ||
| 41 | TERF1 | 14293 | 7219 | 0.005 | 0.3918 | No | ||
| 42 | PIGT | 8134 1 | 7552 | 0.005 | 0.3743 | No | ||
| 43 | CHES1 | 21007 11200 | 7604 | 0.004 | 0.3720 | No | ||
| 44 | PPP2R2A | 3222 21774 | 7796 | 0.004 | 0.3620 | No | ||
| 45 | OTOR | 14825 | 8619 | 0.002 | 0.3179 | No | ||
| 46 | EVI1 | 4689 8926 | 8757 | 0.002 | 0.3107 | No | ||
| 47 | TESK2 | 10504 | 8796 | 0.002 | 0.3088 | No | ||
| 48 | FOXP2 | 4312 17523 8523 | 8879 | 0.002 | 0.3046 | No | ||
| 49 | ARVCF | 8629 | 8919 | 0.002 | 0.3026 | No | ||
| 50 | LMO1 | 17685 | 9048 | 0.001 | 0.2959 | No | ||
| 51 | NR3C2 | 18562 13798 | 9628 | 0.000 | 0.2646 | No | ||
| 52 | SP2 | 8130 | 9663 | 0.000 | 0.2628 | No | ||
| 53 | FIP1L1 | 888 16826 | 10098 | -0.001 | 0.2395 | No | ||
| 54 | ANGPTL1 | 10195 8589 13064 | 10438 | -0.001 | 0.2213 | No | ||
| 55 | LDB2 | 16538 4989 | 10706 | -0.002 | 0.2071 | No | ||
| 56 | ADCY8 | 22281 | 10806 | -0.002 | 0.2019 | No | ||
| 57 | PLK4 | 1870 1874 | 10855 | -0.002 | 0.1995 | No | ||
| 58 | IL4 | 9174 | 11021 | -0.002 | 0.1908 | No | ||
| 59 | CLDN16 | 22802 | 11531 | -0.003 | 0.1637 | No | ||
| 60 | KPNB1 | 20274 | 11790 | -0.004 | 0.1502 | No | ||
| 61 | CACNG8 | 13498 | 11929 | -0.004 | 0.1431 | No | ||
| 62 | RBL2 | 18530 3781 | 12095 | -0.005 | 0.1347 | No | ||
| 63 | CLDN14 | 12081 | 12156 | -0.005 | 0.1319 | No | ||
| 64 | HOXA6 | 17150 | 12169 | -0.005 | 0.1317 | No | ||
| 65 | ALDH1A2 | 19386 | 12280 | -0.005 | 0.1263 | No | ||
| 66 | ARHGAP5 | 4412 8625 | 12380 | -0.005 | 0.1215 | No | ||
| 67 | ATOH1 | 4419 | 12787 | -0.006 | 0.1002 | No | ||
| 68 | MYH14 | 17845 | 13672 | -0.010 | 0.0534 | No | ||
| 69 | GGN | 2472 1171 18312 | 14042 | -0.011 | 0.0346 | No | ||
| 70 | TBX19 | 13780 | 14136 | -0.012 | 0.0308 | No | ||
| 71 | NRXN1 | 1581 1575 22875 | 14207 | -0.012 | 0.0282 | No | ||
| 72 | SLC26A9 | 11560 980 4067 | 14859 | -0.018 | -0.0052 | No | ||
| 73 | SNX26 | 10582 | 14999 | -0.019 | -0.0108 | No | ||
| 74 | PPM1L | 10811 6316 | 15000 | -0.020 | -0.0089 | No | ||
| 75 | GRIA3 | 24349 11996 2649 | 15362 | -0.025 | -0.0259 | No | ||
| 76 | MTMR4 | 20712 | 15520 | -0.029 | -0.0316 | No | ||
| 77 | UFD1L | 22825 | 15711 | -0.035 | -0.0385 | No | ||
| 78 | PHF12 | 11192 6483 | 15889 | -0.041 | -0.0440 | No | ||
| 79 | PPP2R5D | 962 1523 22957 10139 | 16199 | -0.056 | -0.0553 | No | ||
| 80 | MKNK2 | 3299 9392 | 16718 | -0.094 | -0.0740 | No | ||
| 81 | PPM1B | 5280 1567 5281 | 16930 | -0.116 | -0.0740 | No | ||
| 82 | RBMS1 | 14580 | 16958 | -0.120 | -0.0637 | No | ||
| 83 | CDK2 | 3438 3373 19592 3322 | 17164 | -0.148 | -0.0604 | No | ||
| 84 | VGLL4 | 10558 | 17425 | -0.191 | -0.0557 | No | ||
| 85 | APRIN | 4145 | 17811 | -0.275 | -0.0497 | No | ||
| 86 | JUN | 15832 | 18245 | -0.472 | -0.0269 | No | ||
| 87 | BZRAP1 | 9882 | 18257 | -0.480 | 0.0194 | No |