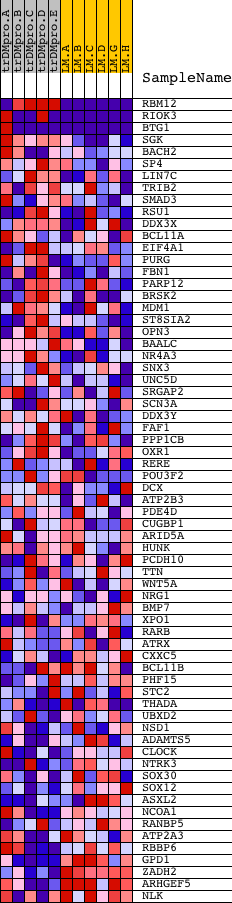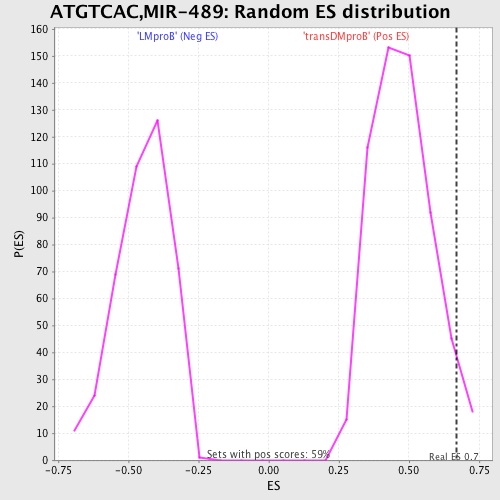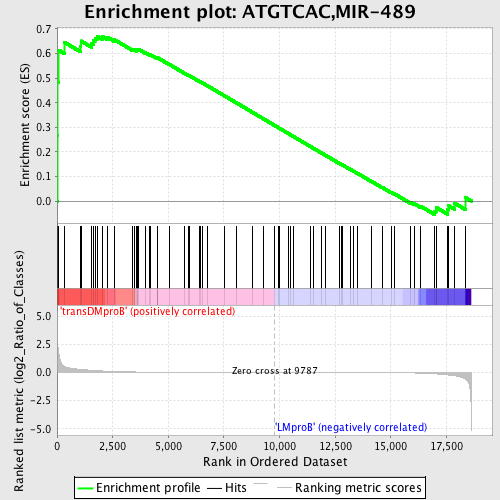
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | ATGTCAC,MIR-489 |
| Enrichment Score (ES) | 0.6709804 |
| Normalized Enrichment Score (NES) | 1.4072447 |
| Nominal p-value | 0.037351444 |
| FDR q-value | 0.57682544 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RBM12 | 7980 2710 13280 | 14 | 3.158 | 0.2665 | Yes | ||
| 2 | RIOK3 | 7354 12425 1973 | 19 | 2.568 | 0.4837 | Yes | ||
| 3 | BTG1 | 4458 4457 8663 | 78 | 1.569 | 0.6134 | Yes | ||
| 4 | SGK | 9809 3419 | 321 | 0.544 | 0.6464 | Yes | ||
| 5 | BACH2 | 8648 | 1042 | 0.269 | 0.6304 | Yes | ||
| 6 | SP4 | 20972 | 1073 | 0.265 | 0.6512 | Yes | ||
| 7 | LIN7C | 14925 | 1545 | 0.190 | 0.6419 | Yes | ||
| 8 | TRIB2 | 21102 | 1630 | 0.176 | 0.6522 | Yes | ||
| 9 | SMAD3 | 19084 | 1706 | 0.168 | 0.6624 | Yes | ||
| 10 | RSU1 | 14683 | 1796 | 0.158 | 0.6710 | Yes | ||
| 11 | DDX3X | 24379 | 2025 | 0.130 | 0.6697 | No | ||
| 12 | BCL11A | 4691 | 2248 | 0.109 | 0.6670 | No | ||
| 13 | EIF4A1 | 8889 23719 | 2569 | 0.084 | 0.6568 | No | ||
| 14 | PURG | 7943 | 3396 | 0.042 | 0.6159 | No | ||
| 15 | FBN1 | 14449 | 3473 | 0.040 | 0.6152 | No | ||
| 16 | PARP12 | 6335 | 3585 | 0.037 | 0.6123 | No | ||
| 17 | BRSK2 | 190 | 3618 | 0.036 | 0.6136 | No | ||
| 18 | MDM1 | 3306 3370 3316 3405 3376 19870 3429 3398 3406 3428 3346 355 | 3626 | 0.035 | 0.6162 | No | ||
| 19 | ST8SIA2 | 17797 | 3640 | 0.035 | 0.6185 | No | ||
| 20 | OPN3 | 13740 | 3969 | 0.028 | 0.6032 | No | ||
| 21 | BAALC | 4407 2201 | 4149 | 0.025 | 0.5956 | No | ||
| 22 | NR4A3 | 9473 16212 5183 | 4183 | 0.024 | 0.5959 | No | ||
| 23 | SNX3 | 20044 3416 | 4489 | 0.020 | 0.5811 | No | ||
| 24 | UNC5D | 9980 | 4519 | 0.020 | 0.5812 | No | ||
| 25 | SRGAP2 | 13836 17045 6433 17046 | 4524 | 0.020 | 0.5827 | No | ||
| 26 | SCN3A | 14573 | 5036 | 0.015 | 0.5564 | No | ||
| 27 | DDX3Y | 6507 4593 | 5744 | 0.011 | 0.5192 | No | ||
| 28 | FAF1 | 2508 16137 8951 | 5889 | 0.010 | 0.5123 | No | ||
| 29 | PPP1CB | 5282 3529 3504 | 5932 | 0.010 | 0.5108 | No | ||
| 30 | OXR1 | 9300 2271 | 6409 | 0.008 | 0.4858 | No | ||
| 31 | RERE | 12722 | 6454 | 0.008 | 0.4841 | No | ||
| 32 | POU3F2 | 15931 | 6528 | 0.007 | 0.4808 | No | ||
| 33 | DCX | 24040 8841 4604 | 6748 | 0.007 | 0.4696 | No | ||
| 34 | ATP2B3 | 24308 11557 | 7537 | 0.005 | 0.4275 | No | ||
| 35 | PDE4D | 10722 6235 | 8054 | 0.003 | 0.4000 | No | ||
| 36 | CUGBP1 | 2805 8819 4576 2924 | 8795 | 0.002 | 0.3602 | No | ||
| 37 | ARID5A | 5668 | 9266 | 0.001 | 0.3350 | No | ||
| 38 | HUNK | 22708 | 9752 | 0.000 | 0.3088 | No | ||
| 39 | PCDH10 | 5227 9534 1893 | 9958 | -0.000 | 0.2978 | No | ||
| 40 | TTN | 14552 2743 14553 | 9975 | -0.000 | 0.2970 | No | ||
| 41 | WNT5A | 22066 5880 | 10412 | -0.001 | 0.2736 | No | ||
| 42 | NRG1 | 3885 | 10499 | -0.001 | 0.2691 | No | ||
| 43 | BMP7 | 14324 | 10620 | -0.002 | 0.2627 | No | ||
| 44 | XPO1 | 4172 | 11397 | -0.003 | 0.2212 | No | ||
| 45 | RARB | 3011 21915 | 11529 | -0.003 | 0.2144 | No | ||
| 46 | ATRX | 10350 5922 2628 | 11881 | -0.004 | 0.1958 | No | ||
| 47 | CXXC5 | 12523 | 12063 | -0.005 | 0.1864 | No | ||
| 48 | BCL11B | 7190 | 12701 | -0.006 | 0.1526 | No | ||
| 49 | PHF15 | 20467 8050 | 12795 | -0.006 | 0.1482 | No | ||
| 50 | STC2 | 5526 | 12846 | -0.007 | 0.1460 | No | ||
| 51 | THADA | 11614 | 13199 | -0.008 | 0.1277 | No | ||
| 52 | UBXD2 | 14150 | 13317 | -0.008 | 0.1221 | No | ||
| 53 | NSD1 | 2134 5197 | 13515 | -0.009 | 0.1122 | No | ||
| 54 | ADAMTS5 | 6207 | 14130 | -0.012 | 0.0801 | No | ||
| 55 | CLOCK | 8749 16507 4530 | 14625 | -0.015 | 0.0548 | No | ||
| 56 | NTRK3 | 2069 3652 3705 9492 | 15029 | -0.020 | 0.0348 | No | ||
| 57 | SOX30 | 1477 20916 | 15172 | -0.022 | 0.0290 | No | ||
| 58 | SOX12 | 5478 | 15876 | -0.040 | -0.0055 | No | ||
| 59 | ASXL2 | 7957 | 16066 | -0.049 | -0.0116 | No | ||
| 60 | NCOA1 | 5154 | 16347 | -0.065 | -0.0212 | No | ||
| 61 | RANBP5 | 21930 | 16967 | -0.122 | -0.0443 | No | ||
| 62 | ATP2A3 | 20795 1314 | 17031 | -0.129 | -0.0367 | No | ||
| 63 | RBBP6 | 5365 | 17059 | -0.133 | -0.0269 | No | ||
| 64 | GPD1 | 22361 | 17548 | -0.217 | -0.0349 | No | ||
| 65 | ZADH2 | 23501 | 17577 | -0.222 | -0.0176 | No | ||
| 66 | ARHGEF5 | 12025 | 17865 | -0.291 | -0.0085 | No | ||
| 67 | NLK | 5179 5178 | 18353 | -0.578 | 0.0142 | No |