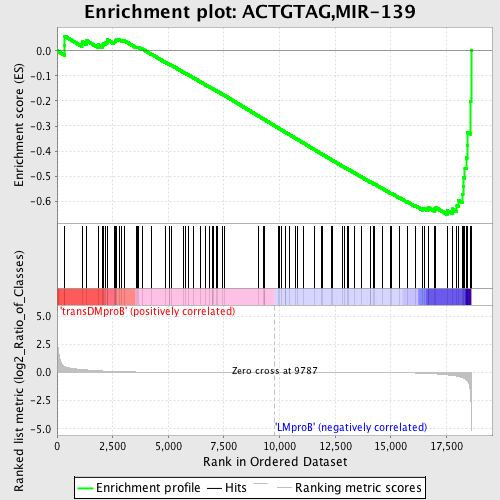
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | ACTGTAG,MIR-139 |
| Enrichment Score (ES) | -0.6526307 |
| Normalized Enrichment Score (NES) | -1.5097624 |
| Nominal p-value | 0.01604278 |
| FDR q-value | 0.5982401 |
| FWER p-Value | 0.911 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSME4 | 8339 20934 | 339 | 0.533 | 0.0204 | No | ||
| 2 | NOTCH1 | 14649 | 347 | 0.525 | 0.0582 | No | ||
| 3 | DHX15 | 8842 | 1119 | 0.255 | 0.0351 | No | ||
| 4 | KBTBD2 | 17137 | 1327 | 0.221 | 0.0400 | No | ||
| 5 | ZFAND3 | 23306 | 1845 | 0.152 | 0.0231 | No | ||
| 6 | DDX3X | 24379 | 2025 | 0.130 | 0.0229 | No | ||
| 7 | DMD | 24295 2647 | 2070 | 0.125 | 0.0296 | No | ||
| 8 | ARRDC3 | 4191 | 2189 | 0.114 | 0.0316 | No | ||
| 9 | MGAT4A | 13977 | 2245 | 0.109 | 0.0365 | No | ||
| 10 | JDP2 | 21201 | 2257 | 0.107 | 0.0437 | No | ||
| 11 | AP3M1 | 7059 | 2557 | 0.085 | 0.0338 | No | ||
| 12 | STAU2 | 6615 | 2603 | 0.082 | 0.0373 | No | ||
| 13 | ZBTB10 | 10468 | 2613 | 0.081 | 0.0427 | No | ||
| 14 | SLC25A3 | 19644 | 2673 | 0.076 | 0.0451 | No | ||
| 15 | CHD7 | 16278 | 2786 | 0.069 | 0.0441 | No | ||
| 16 | HMGCR | 9097 | 2912 | 0.063 | 0.0419 | No | ||
| 17 | EIF4G2 | 1908 8892 | 3012 | 0.058 | 0.0408 | No | ||
| 18 | RSBN1L | 16598 | 3562 | 0.038 | 0.0139 | No | ||
| 19 | SOX5 | 9850 1044 5480 16937 | 3612 | 0.036 | 0.0138 | No | ||
| 20 | AZIN1 | 22307 | 3658 | 0.035 | 0.0139 | No | ||
| 21 | HOXA9 | 17147 9109 1024 | 3829 | 0.031 | 0.0070 | No | ||
| 22 | BTG3 | 8664 | 4257 | 0.023 | -0.0143 | No | ||
| 23 | AEBP2 | 4359 | 4852 | 0.016 | -0.0452 | No | ||
| 24 | AP1S2 | 8423 4228 | 5069 | 0.015 | -0.0558 | No | ||
| 25 | FREM1 | 141 | 5073 | 0.015 | -0.0549 | No | ||
| 26 | MNT | 20784 | 5154 | 0.014 | -0.0582 | No | ||
| 27 | PTPRU | 15738 | 5683 | 0.011 | -0.0859 | No | ||
| 28 | XPO4 | 7161 | 5755 | 0.011 | -0.0890 | No | ||
| 29 | GRM1 | 19822 | 5893 | 0.010 | -0.0957 | No | ||
| 30 | PPARGC1A | 16533 | 6148 | 0.009 | -0.1087 | No | ||
| 31 | LRCH2 | 24035 | 6439 | 0.008 | -0.1238 | No | ||
| 32 | NXPH1 | 17524 | 6686 | 0.007 | -0.1366 | No | ||
| 33 | FBXL16 | 10071 | 6829 | 0.007 | -0.1438 | No | ||
| 34 | PHF6 | 2648 24340 | 6975 | 0.006 | -0.1512 | No | ||
| 35 | PRPF4B | 3208 5295 | 7003 | 0.006 | -0.1522 | No | ||
| 36 | PMP22 | 9594 1312 | 7046 | 0.006 | -0.1540 | No | ||
| 37 | TSPAN3 | 3132 19105 | 7181 | 0.006 | -0.1609 | No | ||
| 38 | TSPAN5 | 15402 | 7212 | 0.005 | -0.1621 | No | ||
| 39 | ETS1 | 10715 6230 3135 | 7418 | 0.005 | -0.1728 | No | ||
| 40 | ANK2 | 4275 | 7519 | 0.005 | -0.1779 | No | ||
| 41 | DNAJC5 | 4568 | 9067 | 0.001 | -0.2613 | No | ||
| 42 | GALNT3 | 4751 | 9286 | 0.001 | -0.2730 | No | ||
| 43 | FBXL7 | 6691 | 9328 | 0.001 | -0.2751 | No | ||
| 44 | SOX1 | 18685 | 9337 | 0.001 | -0.2755 | No | ||
| 45 | PCDH10 | 5227 9534 1893 | 9958 | -0.000 | -0.3089 | No | ||
| 46 | DCBLD2 | 7855 | 9999 | -0.000 | -0.3111 | No | ||
| 47 | FBN2 | 23433 | 10070 | -0.001 | -0.3148 | No | ||
| 48 | SCHIP1 | 15569 | 10260 | -0.001 | -0.3249 | No | ||
| 49 | PPP3CB | 5285 | 10274 | -0.001 | -0.3256 | No | ||
| 50 | CDC42 | 4503 8722 4504 2465 | 10446 | -0.001 | -0.3347 | No | ||
| 51 | ATP2B2 | 17038 | 10695 | -0.002 | -0.3480 | No | ||
| 52 | GDF10 | 4767 | 10817 | -0.002 | -0.3544 | No | ||
| 53 | SSX2IP | 13617 | 11058 | -0.002 | -0.3671 | No | ||
| 54 | TBX1 | 10039 5630 | 11554 | -0.003 | -0.3936 | No | ||
| 55 | ATRX | 10350 5922 2628 | 11881 | -0.004 | -0.4109 | No | ||
| 56 | NME7 | 926 4101 14070 3986 3946 931 | 11943 | -0.004 | -0.4139 | No | ||
| 57 | FMR1 | 24321 | 12324 | -0.005 | -0.4340 | No | ||
| 58 | NDRG4 | 10626 | 12395 | -0.005 | -0.4374 | No | ||
| 59 | NRK | 6559 | 12818 | -0.007 | -0.4597 | No | ||
| 60 | HIPK1 | 4851 | 12913 | -0.007 | -0.4643 | No | ||
| 61 | NPTX1 | 20126 | 13066 | -0.007 | -0.4720 | No | ||
| 62 | HNRPF | 13607 | 13103 | -0.007 | -0.4734 | No | ||
| 63 | ATXN1 | 21477 | 13356 | -0.008 | -0.4864 | No | ||
| 64 | TROVE2 | 13812 | 13667 | -0.010 | -0.5024 | No | ||
| 65 | PAK7 | 14417 | 14063 | -0.012 | -0.5229 | No | ||
| 66 | CHFR | 3646 16757 3624 | 14095 | -0.012 | -0.5237 | No | ||
| 67 | MAT2A | 10553 10554 6099 | 14240 | -0.013 | -0.5306 | No | ||
| 68 | ESRRG | 6447 | 14246 | -0.013 | -0.5299 | No | ||
| 69 | LRFN5 | 6213 | 14605 | -0.015 | -0.5481 | No | ||
| 70 | PPP2CA | 20890 | 14993 | -0.019 | -0.5676 | No | ||
| 71 | PDE3A | 7045 | 15014 | -0.020 | -0.5673 | No | ||
| 72 | KPNA4 | 15321 | 15411 | -0.026 | -0.5867 | No | ||
| 73 | PDE4A | 3037 19542 3083 3023 | 15728 | -0.035 | -0.6012 | No | ||
| 74 | YWHAQ | 10369 | 16091 | -0.050 | -0.6171 | No | ||
| 75 | YWHAG | 16339 | 16425 | -0.070 | -0.6300 | No | ||
| 76 | CACNA2D2 | 7154 | 16496 | -0.075 | -0.6283 | No | ||
| 77 | FUSIP1 | 4715 16036 | 16675 | -0.091 | -0.6313 | No | ||
| 78 | RTF1 | 14899 | 16708 | -0.093 | -0.6263 | No | ||
| 79 | FRMD4A | 5557 | 16945 | -0.118 | -0.6304 | No | ||
| 80 | HDGF | 9082 4846 4845 | 17027 | -0.129 | -0.6254 | No | ||
| 81 | TCF12 | 10044 3125 | 17532 | -0.213 | -0.6372 | Yes | ||
| 82 | DDIT4 | 19762 | 17763 | -0.261 | -0.6306 | Yes | ||
| 83 | FOXP1 | 4242 | 17971 | -0.329 | -0.6178 | Yes | ||
| 84 | MARCH7 | 7172 2698 | 18044 | -0.357 | -0.5957 | Yes | ||
| 85 | TOP1 | 5790 5789 10210 | 18222 | -0.452 | -0.5724 | Yes | ||
| 86 | JUN | 15832 | 18245 | -0.472 | -0.5393 | Yes | ||
| 87 | APLP2 | 19190 | 18258 | -0.480 | -0.5050 | Yes | ||
| 88 | IVNS1ABP | 4384 4050 | 18332 | -0.558 | -0.4684 | Yes | ||
| 89 | SMARCA4 | 19538 | 18387 | -0.620 | -0.4262 | Yes | ||
| 90 | FOS | 21202 | 18426 | -0.702 | -0.3772 | Yes | ||
| 91 | DTX3 | 19607 | 18446 | -0.729 | -0.3252 | Yes | ||
| 92 | SOCS2 | 5694 | 18575 | -1.790 | -0.2020 | Yes | ||
| 93 | NFAT5 | 3921 7037 12036 | 18604 | -2.808 | 0.0006 | Yes |

