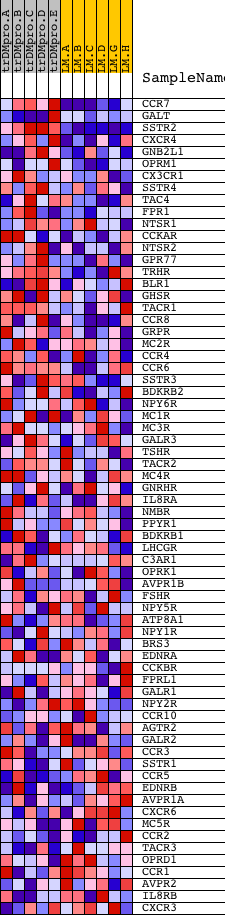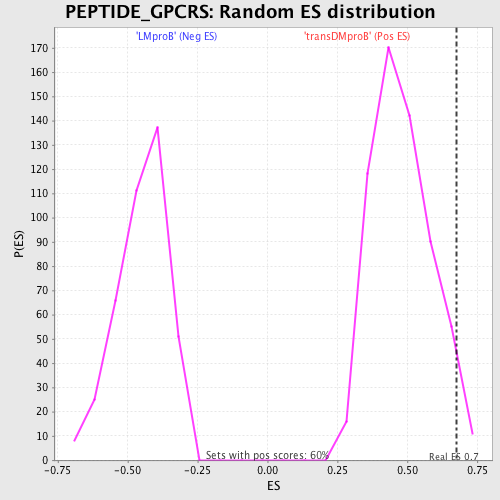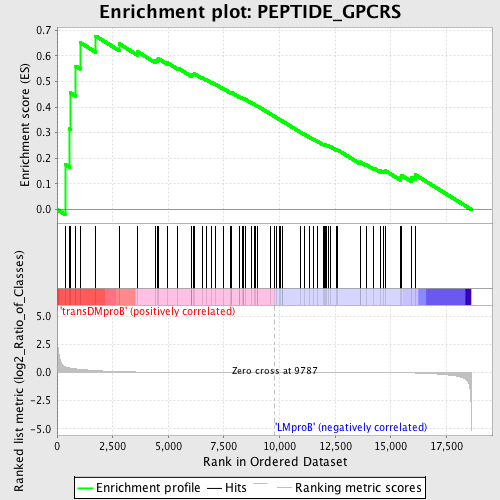
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | PEPTIDE_GPCRS |
| Enrichment Score (ES) | 0.6766228 |
| Normalized Enrichment Score (NES) | 1.4113454 |
| Nominal p-value | 0.03322259 |
| FDR q-value | 0.93241864 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCR7 | 20254 | 359 | 0.510 | 0.1761 | Yes | ||
| 2 | GALT | 16237 2399 | 558 | 0.389 | 0.3145 | Yes | ||
| 3 | SSTR2 | 20609 | 593 | 0.372 | 0.4554 | Yes | ||
| 4 | CXCR4 | 13844 | 842 | 0.307 | 0.5596 | Yes | ||
| 5 | GNB2L1 | 20911 | 1055 | 0.267 | 0.6506 | Yes | ||
| 6 | OPRM1 | 5212 | 1741 | 0.164 | 0.6766 | Yes | ||
| 7 | CX3CR1 | 18965 | 2792 | 0.069 | 0.6465 | No | ||
| 8 | SSTR4 | 9836 | 3620 | 0.036 | 0.6156 | No | ||
| 9 | TAC4 | 20696 | 4428 | 0.021 | 0.5800 | No | ||
| 10 | FPR1 | 23115 | 4501 | 0.020 | 0.5837 | No | ||
| 11 | NTSR1 | 14710 | 4537 | 0.019 | 0.5893 | No | ||
| 12 | CCKAR | 16531 | 4955 | 0.015 | 0.5727 | No | ||
| 13 | NTSR2 | 21310 | 5429 | 0.012 | 0.5520 | No | ||
| 14 | GPR77 | 17958 | 6058 | 0.009 | 0.5217 | No | ||
| 15 | TRHR | 22482 | 6059 | 0.009 | 0.5252 | No | ||
| 16 | BLR1 | 19145 3154 | 6061 | 0.009 | 0.5287 | No | ||
| 17 | GHSR | 1810 15625 | 6134 | 0.009 | 0.5283 | No | ||
| 18 | TACR1 | 1082 17403 | 6172 | 0.009 | 0.5296 | No | ||
| 19 | CCR8 | 19272 | 6518 | 0.008 | 0.5139 | No | ||
| 20 | GRPR | 24015 | 6718 | 0.007 | 0.5058 | No | ||
| 21 | MC2R | 23412 | 6948 | 0.006 | 0.4958 | No | ||
| 22 | CCR4 | 18978 | 7105 | 0.006 | 0.4896 | No | ||
| 23 | CCR6 | 1505 23390 | 7480 | 0.005 | 0.4713 | No | ||
| 24 | SSTR3 | 22218 | 7798 | 0.004 | 0.4557 | No | ||
| 25 | BDKRB2 | 21163 | 7848 | 0.004 | 0.4545 | No | ||
| 26 | NPY6R | 23567 | 7855 | 0.004 | 0.4557 | No | ||
| 27 | MC1R | 18428 | 8178 | 0.003 | 0.4395 | No | ||
| 28 | MC3R | 14721 | 8208 | 0.003 | 0.4391 | No | ||
| 29 | GALR3 | 9000 | 8335 | 0.003 | 0.4333 | No | ||
| 30 | TSHR | 5801 10223 | 8377 | 0.003 | 0.4322 | No | ||
| 31 | TACR2 | 20004 | 8378 | 0.003 | 0.4332 | No | ||
| 32 | MC4R | 13701 | 8447 | 0.003 | 0.4305 | No | ||
| 33 | GNRHR | 4791 | 8733 | 0.002 | 0.4159 | No | ||
| 34 | IL8RA | 13925 | 8871 | 0.002 | 0.4092 | No | ||
| 35 | NMBR | 9466 | 8934 | 0.002 | 0.4065 | No | ||
| 36 | PPYR1 | 21881 | 8992 | 0.001 | 0.4040 | No | ||
| 37 | BDKRB1 | 19418 | 9022 | 0.001 | 0.4029 | No | ||
| 38 | LHCGR | 9273 4994 | 9594 | 0.000 | 0.3723 | No | ||
| 39 | C3AR1 | 8668 | 9769 | 0.000 | 0.3629 | No | ||
| 40 | OPRK1 | 14300 | 9839 | -0.000 | 0.3593 | No | ||
| 41 | AVPR1B | 6441 | 10007 | -0.000 | 0.3504 | No | ||
| 42 | FSHR | 1492 22876 | 10041 | -0.001 | 0.3489 | No | ||
| 43 | NPY5R | 18866 | 10108 | -0.001 | 0.3456 | No | ||
| 44 | ATP8A1 | 4428 | 10110 | -0.001 | 0.3457 | No | ||
| 45 | NPY1R | 9482 | 10935 | -0.002 | 0.3021 | No | ||
| 46 | BRS3 | 24333 | 11113 | -0.003 | 0.2936 | No | ||
| 47 | EDNRA | 18834 | 11343 | -0.003 | 0.2824 | No | ||
| 48 | CCKBR | 8706 | 11507 | -0.003 | 0.2748 | No | ||
| 49 | FPRL1 | 8982 | 11717 | -0.004 | 0.2650 | No | ||
| 50 | GALR1 | 23397 | 11953 | -0.004 | 0.2540 | No | ||
| 51 | NPY2R | 343 | 12018 | -0.004 | 0.2523 | No | ||
| 52 | CCR10 | 20218 | 12042 | -0.005 | 0.2528 | No | ||
| 53 | AGTR2 | 24365 | 12114 | -0.005 | 0.2507 | No | ||
| 54 | GALR2 | 20587 | 12192 | -0.005 | 0.2485 | No | ||
| 55 | CCR3 | 19251 | 12296 | -0.005 | 0.2449 | No | ||
| 56 | SSTR1 | 21264 | 12548 | -0.006 | 0.2335 | No | ||
| 57 | CCR5 | 8754 4534 | 12615 | -0.006 | 0.2323 | No | ||
| 58 | EDNRB | 21728 3564 | 13614 | -0.009 | 0.1821 | No | ||
| 59 | AVPR1A | 19863 | 13617 | -0.009 | 0.1856 | No | ||
| 60 | CXCR6 | 19252 | 13897 | -0.011 | 0.1746 | No | ||
| 61 | MC5R | 23524 | 14227 | -0.013 | 0.1617 | No | ||
| 62 | CCR2 | 19250 | 14534 | -0.015 | 0.1507 | No | ||
| 63 | TACR3 | 15415 | 14675 | -0.016 | 0.1492 | No | ||
| 64 | OPRD1 | 15737 | 14757 | -0.016 | 0.1512 | No | ||
| 65 | CCR1 | 18951 | 15449 | -0.027 | 0.1243 | No | ||
| 66 | AVPR2 | 8646 | 15466 | -0.027 | 0.1339 | No | ||
| 67 | IL8RB | 8752 4533 | 15940 | -0.043 | 0.1249 | No | ||
| 68 | CXCR3 | 24093 | 16094 | -0.050 | 0.1360 | No |