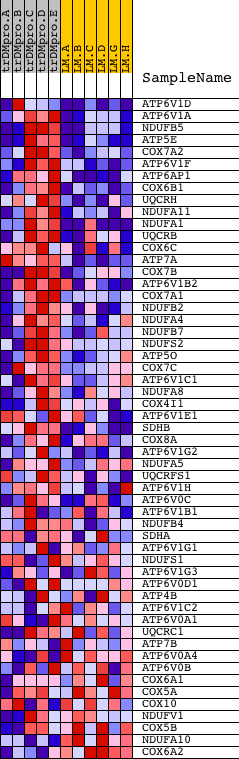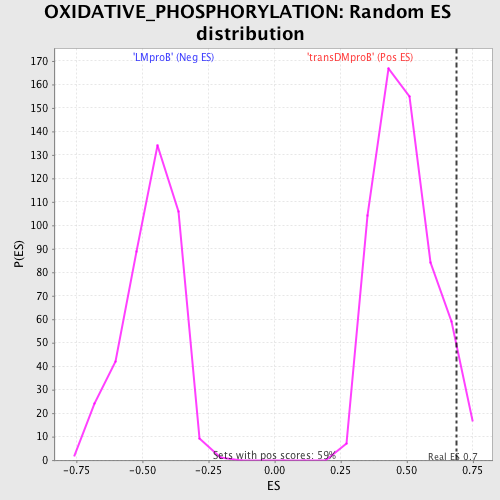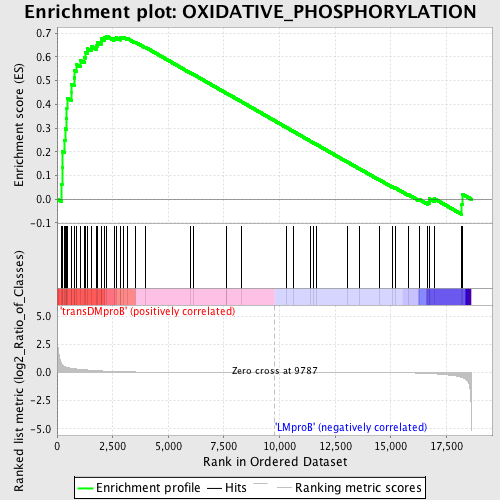
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | 0.6872689 |
| Normalized Enrichment Score (NES) | 1.3995812 |
| Nominal p-value | 0.047217537 |
| FDR q-value | 0.7398737 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP6V1D | 7872 | 208 | 0.750 | 0.0627 | Yes | ||
| 2 | ATP6V1A | 8638 | 223 | 0.709 | 0.1319 | Yes | ||
| 3 | NDUFB5 | 12277 | 229 | 0.698 | 0.2006 | Yes | ||
| 4 | ATP5E | 14321 | 326 | 0.541 | 0.2487 | Yes | ||
| 5 | COX7A2 | 19053 | 361 | 0.508 | 0.2970 | Yes | ||
| 6 | ATP6V1F | 12291 | 419 | 0.462 | 0.3395 | Yes | ||
| 7 | ATP6AP1 | 24296 | 440 | 0.449 | 0.3828 | Yes | ||
| 8 | COX6B1 | 4283 8479 | 445 | 0.445 | 0.4265 | Yes | ||
| 9 | UQCRH | 12378 | 654 | 0.356 | 0.4503 | Yes | ||
| 10 | NDUFA11 | 12832 | 657 | 0.354 | 0.4852 | Yes | ||
| 11 | NDUFA1 | 24170 | 765 | 0.323 | 0.5113 | Yes | ||
| 12 | UQCRB | 12545 | 779 | 0.320 | 0.5421 | Yes | ||
| 13 | COX6C | 8775 | 873 | 0.300 | 0.5667 | Yes | ||
| 14 | ATP7A | 4425 | 1037 | 0.271 | 0.5846 | Yes | ||
| 15 | COX7B | 7260 | 1233 | 0.233 | 0.5971 | Yes | ||
| 16 | ATP6V1B2 | 18599 | 1276 | 0.228 | 0.6173 | Yes | ||
| 17 | COX7A1 | 4554 | 1359 | 0.217 | 0.6343 | Yes | ||
| 18 | NDUFB2 | 7542 | 1536 | 0.191 | 0.6437 | Yes | ||
| 19 | NDUFA4 | 9451 | 1786 | 0.159 | 0.6459 | Yes | ||
| 20 | NDUFB7 | 12429 | 1797 | 0.158 | 0.6609 | Yes | ||
| 21 | NDUFS2 | 4089 13760 | 1978 | 0.135 | 0.6645 | Yes | ||
| 22 | ATP5O | 22539 | 1996 | 0.133 | 0.6768 | Yes | ||
| 23 | COX7C | 8776 | 2137 | 0.118 | 0.6809 | Yes | ||
| 24 | ATP6V1C1 | 22487 12329 | 2223 | 0.111 | 0.6873 | Yes | ||
| 25 | NDUFA8 | 14605 | 2559 | 0.085 | 0.6776 | No | ||
| 26 | COX4I1 | 18444 | 2649 | 0.079 | 0.6805 | No | ||
| 27 | ATP6V1E1 | 4423 | 2830 | 0.067 | 0.6774 | No | ||
| 28 | SDHB | 2348 12566 14880 | 2862 | 0.065 | 0.6822 | No | ||
| 29 | COX8A | 8777 | 2965 | 0.060 | 0.6827 | No | ||
| 30 | ATP6V1G2 | 23257 1534 | 3166 | 0.051 | 0.6769 | No | ||
| 31 | NDUFA5 | 1076 7544 | 3509 | 0.039 | 0.6623 | No | ||
| 32 | UQCRFS1 | 21499 | 3972 | 0.028 | 0.6401 | No | ||
| 33 | ATP6V1H | 14301 | 5985 | 0.010 | 0.5327 | No | ||
| 34 | ATP6V0C | 8643 | 6133 | 0.009 | 0.5256 | No | ||
| 35 | ATP6V1B1 | 8499 | 7594 | 0.004 | 0.4474 | No | ||
| 36 | NDUFB4 | 22596 7541 | 7629 | 0.004 | 0.4460 | No | ||
| 37 | SDHA | 12436 | 8271 | 0.003 | 0.4118 | No | ||
| 38 | ATP6V1G1 | 12322 | 10293 | -0.001 | 0.3030 | No | ||
| 39 | NDUFS1 | 13940 | 10638 | -0.002 | 0.2846 | No | ||
| 40 | ATP6V1G3 | 14107 | 11395 | -0.003 | 0.2442 | No | ||
| 41 | ATP6V0D1 | 3895 8639 3920 | 11536 | -0.003 | 0.2370 | No | ||
| 42 | ATP4B | 18928 | 11636 | -0.004 | 0.2320 | No | ||
| 43 | ATP6V1C2 | 21098 | 13041 | -0.007 | 0.1571 | No | ||
| 44 | ATP6V0A1 | 8640 4424 1197 | 13577 | -0.009 | 0.1292 | No | ||
| 45 | UQCRC1 | 10259 | 14484 | -0.014 | 0.0818 | No | ||
| 46 | ATP7B | 18918 4426 4427 8641 | 15054 | -0.020 | 0.0531 | No | ||
| 47 | ATP6V0A4 | 8934 | 15200 | -0.022 | 0.0475 | No | ||
| 48 | ATP6V0B | 15786 | 15777 | -0.036 | 0.0201 | No | ||
| 49 | COX6A1 | 8774 4553 | 16268 | -0.060 | -0.0004 | No | ||
| 50 | COX5A | 19431 | 16647 | -0.088 | -0.0121 | No | ||
| 51 | COX10 | 20408 | 16725 | -0.095 | -0.0069 | No | ||
| 52 | NDUFV1 | 23953 | 16742 | -0.097 | 0.0018 | No | ||
| 53 | COX5B | 4552 | 16962 | -0.121 | 0.0020 | No | ||
| 54 | NDUFA10 | 7420 | 18192 | -0.435 | -0.0214 | No | ||
| 55 | COX6A2 | 17605 | 18215 | -0.448 | 0.0216 | No |