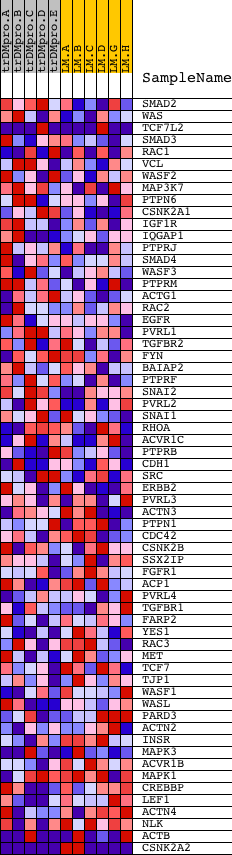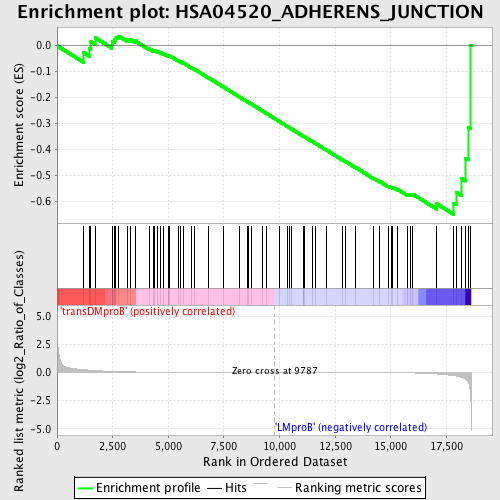
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04520_ADHERENS_JUNCTION |
| Enrichment Score (ES) | -0.6509649 |
| Normalized Enrichment Score (NES) | -1.4075109 |
| Nominal p-value | 0.03931204 |
| FDR q-value | 0.76471364 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SMAD2 | 23511 | 1181 | 0.244 | -0.0264 | No | ||
| 2 | WAS | 24193 | 1467 | 0.201 | -0.0110 | No | ||
| 3 | TCF7L2 | 10048 5646 | 1520 | 0.193 | 0.0157 | No | ||
| 4 | SMAD3 | 19084 | 1706 | 0.168 | 0.0315 | No | ||
| 5 | RAC1 | 16302 | 2467 | 0.090 | 0.0043 | No | ||
| 6 | VCL | 22083 | 2490 | 0.088 | 0.0167 | No | ||
| 7 | WASF2 | 6326 | 2591 | 0.083 | 0.0240 | No | ||
| 8 | MAP3K7 | 16255 | 2645 | 0.079 | 0.0332 | No | ||
| 9 | PTPN6 | 17002 | 2774 | 0.070 | 0.0370 | No | ||
| 10 | CSNK2A1 | 14797 | 3151 | 0.051 | 0.0246 | No | ||
| 11 | IGF1R | 9157 | 3281 | 0.046 | 0.0248 | No | ||
| 12 | IQGAP1 | 6619 | 3500 | 0.039 | 0.0190 | No | ||
| 13 | PTPRJ | 9664 | 4130 | 0.025 | -0.0111 | No | ||
| 14 | SMAD4 | 5058 | 4313 | 0.022 | -0.0175 | No | ||
| 15 | WASF3 | 16625 | 4387 | 0.021 | -0.0182 | No | ||
| 16 | PTPRM | 22904 | 4507 | 0.020 | -0.0216 | No | ||
| 17 | ACTG1 | 8535 | 4653 | 0.018 | -0.0266 | No | ||
| 18 | RAC2 | 22217 | 4779 | 0.017 | -0.0308 | No | ||
| 19 | EGFR | 1329 20944 | 4992 | 0.015 | -0.0399 | No | ||
| 20 | PVRL1 | 3006 19482 12211 7192 | 5015 | 0.015 | -0.0387 | No | ||
| 21 | TGFBR2 | 2994 3001 10167 | 5038 | 0.015 | -0.0377 | No | ||
| 22 | FYN | 3375 3395 20052 | 5437 | 0.012 | -0.0572 | No | ||
| 23 | BAIAP2 | 449 4236 | 5566 | 0.012 | -0.0624 | No | ||
| 24 | PTPRF | 5331 2545 | 5661 | 0.011 | -0.0657 | No | ||
| 25 | SNAI2 | 22848 | 6051 | 0.009 | -0.0853 | No | ||
| 26 | PVRL2 | 9671 5336 | 6192 | 0.009 | -0.0915 | No | ||
| 27 | SNAI1 | 14726 | 6823 | 0.007 | -0.1244 | No | ||
| 28 | RHOA | 8624 4409 4410 | 7465 | 0.005 | -0.1583 | No | ||
| 29 | ACVR1C | 14585 | 8205 | 0.003 | -0.1976 | No | ||
| 30 | PTPRB | 19875 | 8579 | 0.002 | -0.2174 | No | ||
| 31 | CDH1 | 18479 | 8600 | 0.002 | -0.2181 | No | ||
| 32 | SRC | 5507 | 8725 | 0.002 | -0.2245 | No | ||
| 33 | ERBB2 | 8913 | 9244 | 0.001 | -0.2522 | No | ||
| 34 | PVRL3 | 1674 22579 1675 | 9397 | 0.001 | -0.2603 | No | ||
| 35 | ACTN3 | 23967 8540 | 9983 | -0.000 | -0.2918 | No | ||
| 36 | PTPN1 | 5325 | 10357 | -0.001 | -0.3117 | No | ||
| 37 | CDC42 | 4503 8722 4504 2465 | 10446 | -0.001 | -0.3163 | No | ||
| 38 | CSNK2B | 23008 | 10540 | -0.001 | -0.3211 | No | ||
| 39 | SSX2IP | 13617 | 11058 | -0.002 | -0.3486 | No | ||
| 40 | FGFR1 | 3789 8968 | 11060 | -0.002 | -0.3483 | No | ||
| 41 | ACP1 | 4317 | 11107 | -0.003 | -0.3503 | No | ||
| 42 | PVRL4 | 14055 970 | 11460 | -0.003 | -0.3688 | No | ||
| 43 | TGFBR1 | 16213 10165 5745 | 11612 | -0.004 | -0.3764 | No | ||
| 44 | FARP2 | 14175 | 12119 | -0.005 | -0.4030 | No | ||
| 45 | YES1 | 5930 | 12834 | -0.007 | -0.4404 | No | ||
| 46 | RAC3 | 20561 | 12968 | -0.007 | -0.4465 | No | ||
| 47 | MET | 17520 | 13399 | -0.008 | -0.4684 | No | ||
| 48 | TCF7 | 1467 20466 | 14220 | -0.012 | -0.5107 | No | ||
| 49 | TJP1 | 17803 | 14501 | -0.014 | -0.5236 | No | ||
| 50 | WASF1 | 13514 | 14885 | -0.018 | -0.5415 | No | ||
| 51 | WASL | 17203 | 15021 | -0.020 | -0.5457 | No | ||
| 52 | PARD3 | 8212 13553 | 15086 | -0.021 | -0.5460 | No | ||
| 53 | ACTN2 | 21545 | 15303 | -0.024 | -0.5540 | No | ||
| 54 | INSR | 18950 | 15741 | -0.035 | -0.5721 | No | ||
| 55 | MAPK3 | 6458 11170 | 15861 | -0.039 | -0.5725 | No | ||
| 56 | ACVR1B | 4335 22353 | 15980 | -0.045 | -0.5720 | No | ||
| 57 | MAPK1 | 1642 11167 | 17069 | -0.135 | -0.6100 | Yes | ||
| 58 | CREBBP | 22682 8783 | 17830 | -0.281 | -0.6080 | Yes | ||
| 59 | LEF1 | 1860 15420 | 17935 | -0.318 | -0.5649 | Yes | ||
| 60 | ACTN4 | 17905 1798 1983 | 18182 | -0.428 | -0.5127 | Yes | ||
| 61 | NLK | 5179 5178 | 18353 | -0.578 | -0.4335 | Yes | ||
| 62 | ACTB | 8534 337 337 338 | 18478 | -0.827 | -0.3136 | Yes | ||
| 63 | CSNK2A2 | 4567 8808 | 18589 | -2.098 | 0.0015 | Yes |