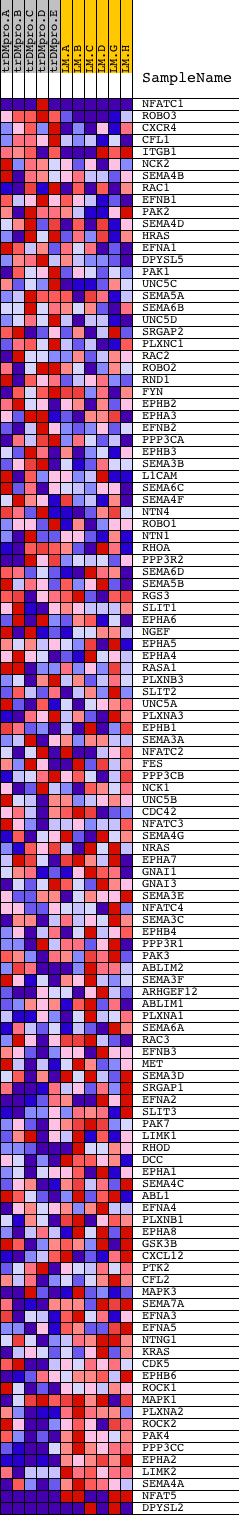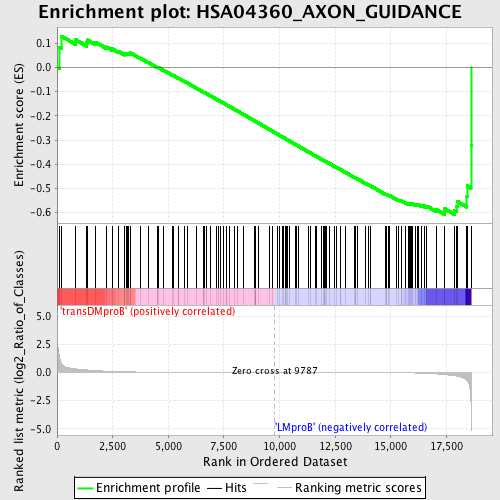
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04360_AXON_GUIDANCE |
| Enrichment Score (ES) | -0.60809606 |
| Normalized Enrichment Score (NES) | -1.4631627 |
| Nominal p-value | 0.0050251256 |
| FDR q-value | 0.6272 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 106 | 1.431 | 0.0847 | No | ||
| 2 | ROBO3 | 9705 | 196 | 0.791 | 0.1298 | No | ||
| 3 | CXCR4 | 13844 | 842 | 0.307 | 0.1143 | No | ||
| 4 | CFL1 | 4516 | 1323 | 0.222 | 0.1024 | No | ||
| 5 | ITGB1 | 3872 18411 | 1364 | 0.217 | 0.1139 | No | ||
| 6 | NCK2 | 9448 | 1716 | 0.167 | 0.1055 | No | ||
| 7 | SEMA4B | 18205 | 2228 | 0.111 | 0.0849 | No | ||
| 8 | RAC1 | 16302 | 2467 | 0.090 | 0.0777 | No | ||
| 9 | EFNB1 | 24285 | 2748 | 0.072 | 0.0671 | No | ||
| 10 | PAK2 | 10310 5875 | 3038 | 0.056 | 0.0551 | No | ||
| 11 | SEMA4D | 9800 | 3040 | 0.056 | 0.0586 | No | ||
| 12 | HRAS | 4868 | 3102 | 0.053 | 0.0586 | No | ||
| 13 | EFNA1 | 15279 | 3169 | 0.050 | 0.0583 | No | ||
| 14 | DPYSL5 | 92 | 3218 | 0.049 | 0.0587 | No | ||
| 15 | PAK1 | 9527 | 3284 | 0.046 | 0.0582 | No | ||
| 16 | UNC5C | 10256 | 3302 | 0.046 | 0.0601 | No | ||
| 17 | SEMA5A | 22496 5423 | 3751 | 0.033 | 0.0380 | No | ||
| 18 | SEMA6B | 1587 22932 | 4087 | 0.026 | 0.0215 | No | ||
| 19 | UNC5D | 9980 | 4519 | 0.020 | -0.0006 | No | ||
| 20 | SRGAP2 | 13836 17045 6433 17046 | 4524 | 0.020 | 0.0005 | No | ||
| 21 | PLXNC1 | 7056 | 4553 | 0.019 | 0.0002 | No | ||
| 22 | RAC2 | 22217 | 4779 | 0.017 | -0.0109 | No | ||
| 23 | ROBO2 | 6501 | 5168 | 0.014 | -0.0310 | No | ||
| 24 | RND1 | 5868 | 5220 | 0.014 | -0.0329 | No | ||
| 25 | FYN | 3375 3395 20052 | 5437 | 0.012 | -0.0438 | No | ||
| 26 | EPHB2 | 4675 2440 8910 | 5459 | 0.012 | -0.0442 | No | ||
| 27 | EPHA3 | 22563 1643 | 5724 | 0.011 | -0.0578 | No | ||
| 28 | EFNB2 | 18938 | 5849 | 0.010 | -0.0638 | No | ||
| 29 | PPP3CA | 1863 5284 | 6266 | 0.008 | -0.0858 | No | ||
| 30 | EPHB3 | 22816 | 6567 | 0.007 | -0.1015 | No | ||
| 31 | SEMA3B | 5422 | 6580 | 0.007 | -0.1017 | No | ||
| 32 | L1CAM | 3 | 6646 | 0.007 | -0.1048 | No | ||
| 33 | SEMA6C | 1797 5424 | 6693 | 0.007 | -0.1068 | No | ||
| 34 | SEMA4F | 17105 | 6882 | 0.006 | -0.1166 | No | ||
| 35 | NTN4 | 19903 | 7143 | 0.006 | -0.1303 | No | ||
| 36 | ROBO1 | 9731 | 7235 | 0.005 | -0.1349 | No | ||
| 37 | NTN1 | 20400 | 7349 | 0.005 | -0.1407 | No | ||
| 38 | RHOA | 8624 4409 4410 | 7465 | 0.005 | -0.1466 | No | ||
| 39 | PPP3R2 | 9612 | 7630 | 0.004 | -0.1552 | No | ||
| 40 | SEMA6D | 2800 14876 90 | 7631 | 0.004 | -0.1549 | No | ||
| 41 | SEMA5B | 9801 22777 | 7750 | 0.004 | -0.1610 | No | ||
| 42 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 7971 | 0.003 | -0.1727 | No | ||
| 43 | SLIT1 | 23678 | 8106 | 0.003 | -0.1797 | No | ||
| 44 | EPHA6 | 22564 | 8361 | 0.003 | -0.1933 | No | ||
| 45 | NGEF | 13891 | 8885 | 0.002 | -0.2215 | No | ||
| 46 | EPHA5 | 16498 | 8927 | 0.002 | -0.2236 | No | ||
| 47 | EPHA4 | 4673 | 8932 | 0.002 | -0.2237 | No | ||
| 48 | RASA1 | 10174 | 8933 | 0.002 | -0.2236 | No | ||
| 49 | PLXNB3 | 24304 | 9051 | 0.001 | -0.2298 | No | ||
| 50 | SLIT2 | 5456 | 9534 | 0.001 | -0.2558 | No | ||
| 51 | UNC5A | 3288 21631 | 9673 | 0.000 | -0.2633 | No | ||
| 52 | PLXNA3 | 24299 | 9887 | -0.000 | -0.2748 | No | ||
| 53 | EPHB1 | 19024 | 9996 | -0.000 | -0.2806 | No | ||
| 54 | SEMA3A | 16919 | 10151 | -0.001 | -0.2889 | No | ||
| 55 | NFATC2 | 5168 2866 | 10189 | -0.001 | -0.2908 | No | ||
| 56 | FES | 17779 3949 | 10245 | -0.001 | -0.2937 | No | ||
| 57 | PPP3CB | 5285 | 10274 | -0.001 | -0.2952 | No | ||
| 58 | NCK1 | 9447 5152 | 10315 | -0.001 | -0.2973 | No | ||
| 59 | UNC5B | 8404 | 10356 | -0.001 | -0.2994 | No | ||
| 60 | CDC42 | 4503 8722 4504 2465 | 10446 | -0.001 | -0.3041 | No | ||
| 61 | NFATC3 | 5169 | 10711 | -0.002 | -0.3183 | No | ||
| 62 | SEMA4G | 11182 | 10762 | -0.002 | -0.3209 | No | ||
| 63 | NRAS | 5191 | 10833 | -0.002 | -0.3245 | No | ||
| 64 | EPHA7 | 8909 4674 | 11298 | -0.003 | -0.3494 | No | ||
| 65 | GNAI1 | 9024 | 11386 | -0.003 | -0.3539 | No | ||
| 66 | GNAI3 | 15198 | 11620 | -0.004 | -0.3663 | No | ||
| 67 | SEMA3E | 16918 | 11650 | -0.004 | -0.3676 | No | ||
| 68 | NFATC4 | 22002 | 11897 | -0.004 | -0.3807 | No | ||
| 69 | SEMA3C | 16914 | 11963 | -0.004 | -0.3839 | No | ||
| 70 | EPHB4 | 4676 | 12011 | -0.004 | -0.3862 | No | ||
| 71 | PPP3R1 | 9611 489 | 12027 | -0.004 | -0.3867 | No | ||
| 72 | PAK3 | 9528 | 12079 | -0.005 | -0.3892 | No | ||
| 73 | ABLIM2 | 16866 3605 | 12096 | -0.005 | -0.3897 | No | ||
| 74 | SEMA3F | 18999 3097 | 12225 | -0.005 | -0.3963 | No | ||
| 75 | ARHGEF12 | 19156 | 12482 | -0.006 | -0.4098 | No | ||
| 76 | ABLIM1 | 3717 3766 3679 | 12537 | -0.006 | -0.4124 | No | ||
| 77 | PLXNA1 | 5268 | 12724 | -0.006 | -0.4220 | No | ||
| 78 | SEMA6A | 23439 9802 | 12748 | -0.006 | -0.4229 | No | ||
| 79 | RAC3 | 20561 | 12968 | -0.007 | -0.4343 | No | ||
| 80 | EFNB3 | 20391 | 13361 | -0.008 | -0.4549 | No | ||
| 81 | MET | 17520 | 13399 | -0.008 | -0.4564 | No | ||
| 82 | SEMA3D | 4240 | 13413 | -0.009 | -0.4566 | No | ||
| 83 | SRGAP1 | 19615 | 13493 | -0.009 | -0.4603 | No | ||
| 84 | EFNA2 | 8883 | 13848 | -0.010 | -0.4788 | No | ||
| 85 | SLIT3 | 20925 | 14010 | -0.011 | -0.4867 | No | ||
| 86 | PAK7 | 14417 | 14063 | -0.012 | -0.4888 | No | ||
| 87 | LIMK1 | 16350 | 14077 | -0.012 | -0.4888 | No | ||
| 88 | RHOD | 23961 | 14763 | -0.017 | -0.5248 | No | ||
| 89 | DCC | 8839 | 14806 | -0.017 | -0.5260 | No | ||
| 90 | EPHA1 | 17168 8908 17167 | 14826 | -0.017 | -0.5259 | No | ||
| 91 | SEMA4C | 9799 | 14897 | -0.018 | -0.5286 | No | ||
| 92 | ABL1 | 2693 4301 2794 | 14958 | -0.019 | -0.5306 | No | ||
| 93 | EFNA4 | 1866 15277 | 15255 | -0.023 | -0.5451 | No | ||
| 94 | PLXNB1 | 19301 | 15363 | -0.025 | -0.5493 | No | ||
| 95 | EPHA8 | 15705 | 15474 | -0.028 | -0.5535 | No | ||
| 96 | GSK3B | 22761 | 15479 | -0.028 | -0.5520 | No | ||
| 97 | CXCL12 | 9792 1182 1031 | 15675 | -0.033 | -0.5605 | No | ||
| 98 | PTK2 | 22271 | 15812 | -0.038 | -0.5654 | No | ||
| 99 | CFL2 | 2155 21069 | 15840 | -0.039 | -0.5644 | No | ||
| 100 | MAPK3 | 6458 11170 | 15861 | -0.039 | -0.5630 | No | ||
| 101 | SEMA7A | 19426 9803 | 15906 | -0.041 | -0.5628 | No | ||
| 102 | EFNA3 | 15278 | 15955 | -0.044 | -0.5626 | No | ||
| 103 | EFNA5 | 4655 8884 | 16100 | -0.051 | -0.5672 | No | ||
| 104 | NTNG1 | 13484 8156 1814 | 16203 | -0.056 | -0.5692 | No | ||
| 105 | KRAS | 9247 | 16247 | -0.058 | -0.5678 | No | ||
| 106 | CDK5 | 16591 | 16392 | -0.068 | -0.5713 | No | ||
| 107 | EPHB6 | 17473 | 16506 | -0.076 | -0.5726 | No | ||
| 108 | ROCK1 | 5386 | 16614 | -0.085 | -0.5730 | No | ||
| 109 | MAPK1 | 1642 11167 | 17069 | -0.135 | -0.5891 | No | ||
| 110 | PLXNA2 | 5269 | 17408 | -0.188 | -0.5955 | Yes | ||
| 111 | ROCK2 | 21309 5387 | 17419 | -0.190 | -0.5840 | Yes | ||
| 112 | PAK4 | 17909 | 17866 | -0.291 | -0.5897 | Yes | ||
| 113 | PPP3CC | 21763 | 17966 | -0.329 | -0.5743 | Yes | ||
| 114 | EPHA2 | 16006 | 17991 | -0.338 | -0.5542 | Yes | ||
| 115 | LIMK2 | 9278 1296 | 18422 | -0.690 | -0.5339 | Yes | ||
| 116 | SEMA4A | 15289 | 18431 | -0.706 | -0.4897 | Yes | ||
| 117 | NFAT5 | 3921 7037 12036 | 18604 | -2.808 | -0.3217 | Yes | ||
| 118 | DPYSL2 | 3122 4561 8787 | 18616 | -5.104 | 0.0000 | Yes |