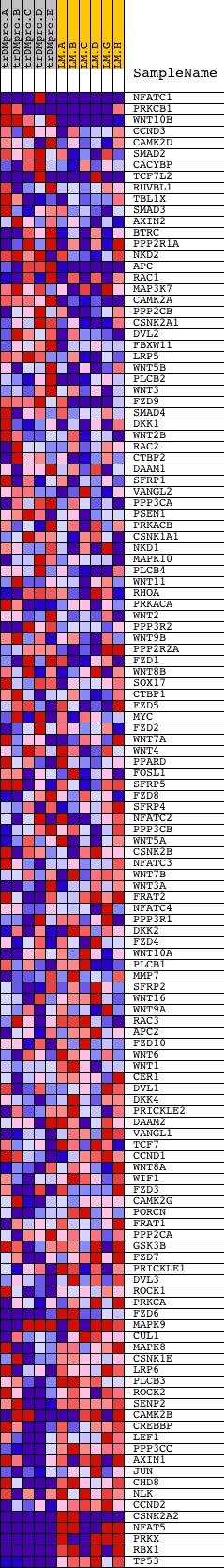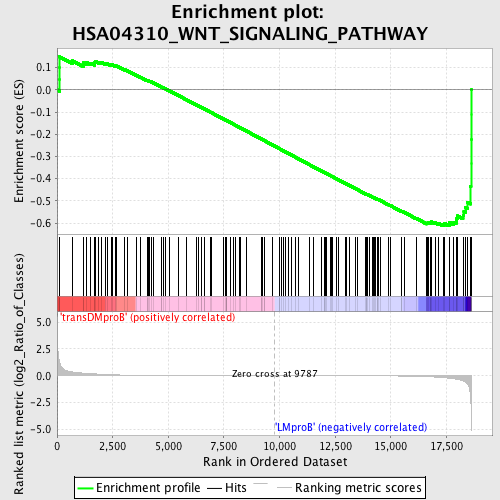
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04310_WNT_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.6131978 |
| Normalized Enrichment Score (NES) | -1.4768853 |
| Nominal p-value | 0.013297873 |
| FDR q-value | 0.6458275 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 106 | 1.431 | 0.0478 | No | ||
| 2 | PRKCB1 | 1693 9574 | 110 | 1.380 | 0.0993 | No | ||
| 3 | WNT10B | 5874 | 114 | 1.331 | 0.1489 | No | ||
| 4 | CCND3 | 4489 4490 | 686 | 0.346 | 0.1309 | No | ||
| 5 | CAMK2D | 4232 | 1178 | 0.244 | 0.1135 | No | ||
| 6 | SMAD2 | 23511 | 1181 | 0.244 | 0.1225 | No | ||
| 7 | CACYBP | 4468 | 1326 | 0.221 | 0.1230 | No | ||
| 8 | TCF7L2 | 10048 5646 | 1520 | 0.193 | 0.1198 | No | ||
| 9 | RUVBL1 | 7131 7130 | 1682 | 0.171 | 0.1174 | No | ||
| 10 | TBL1X | 5628 | 1695 | 0.169 | 0.1231 | No | ||
| 11 | SMAD3 | 19084 | 1706 | 0.168 | 0.1289 | No | ||
| 12 | AXIN2 | 20618 | 1874 | 0.148 | 0.1254 | No | ||
| 13 | BTRC | 4459 | 1991 | 0.134 | 0.1241 | No | ||
| 14 | PPP2R1A | 11951 | 2184 | 0.115 | 0.1180 | No | ||
| 15 | NKD2 | 7798 | 2269 | 0.106 | 0.1174 | No | ||
| 16 | APC | 4396 2022 | 2443 | 0.092 | 0.1115 | No | ||
| 17 | RAC1 | 16302 | 2467 | 0.090 | 0.1137 | No | ||
| 18 | MAP3K7 | 16255 | 2645 | 0.079 | 0.1070 | No | ||
| 19 | CAMK2A | 2024 23541 1980 | 2682 | 0.076 | 0.1079 | No | ||
| 20 | PPP2CB | 18636 | 3031 | 0.057 | 0.0912 | No | ||
| 21 | CSNK2A1 | 14797 | 3151 | 0.051 | 0.0867 | No | ||
| 22 | DVL2 | 20813 | 3584 | 0.037 | 0.0647 | No | ||
| 23 | FBXW11 | 20926 | 3738 | 0.033 | 0.0577 | No | ||
| 24 | LRP5 | 23948 9285 | 4054 | 0.026 | 0.0416 | No | ||
| 25 | WNT5B | 17022 | 4082 | 0.026 | 0.0411 | No | ||
| 26 | PLCB2 | 5262 | 4097 | 0.026 | 0.0413 | No | ||
| 27 | WNT3 | 20635 | 4150 | 0.025 | 0.0394 | No | ||
| 28 | FZD9 | 16345 | 4229 | 0.023 | 0.0361 | No | ||
| 29 | SMAD4 | 5058 | 4313 | 0.022 | 0.0324 | No | ||
| 30 | DKK1 | 23700 | 4692 | 0.018 | 0.0127 | No | ||
| 31 | WNT2B | 15214 1838 | 4713 | 0.018 | 0.0122 | No | ||
| 32 | RAC2 | 22217 | 4779 | 0.017 | 0.0094 | No | ||
| 33 | CTBP2 | 17591 1137 | 4870 | 0.016 | 0.0051 | No | ||
| 34 | DAAM1 | 5536 | 5063 | 0.015 | -0.0047 | No | ||
| 35 | SFRP1 | 9805 | 5457 | 0.012 | -0.0256 | No | ||
| 36 | VANGL2 | 4109 8220 | 5811 | 0.010 | -0.0443 | No | ||
| 37 | PPP3CA | 1863 5284 | 6266 | 0.008 | -0.0685 | No | ||
| 38 | PSEN1 | 5297 2125 9630 | 6343 | 0.008 | -0.0723 | No | ||
| 39 | PRKACB | 15140 | 6469 | 0.008 | -0.0788 | No | ||
| 40 | CSNK1A1 | 8204 | 6510 | 0.008 | -0.0807 | No | ||
| 41 | NKD1 | 8228 | 6605 | 0.007 | -0.0855 | No | ||
| 42 | MAPK10 | 11169 | 6611 | 0.007 | -0.0855 | No | ||
| 43 | PLCB4 | 9586 | 6900 | 0.006 | -0.1008 | No | ||
| 44 | WNT11 | 5876 | 6936 | 0.006 | -0.1025 | No | ||
| 45 | RHOA | 8624 4409 4410 | 7465 | 0.005 | -0.1309 | No | ||
| 46 | PRKACA | 18549 3844 | 7561 | 0.005 | -0.1358 | No | ||
| 47 | WNT2 | 17212 | 7606 | 0.004 | -0.1381 | No | ||
| 48 | PPP3R2 | 9612 | 7630 | 0.004 | -0.1391 | No | ||
| 49 | WNT9B | 20193 | 7635 | 0.004 | -0.1392 | No | ||
| 50 | PPP2R2A | 3222 21774 | 7796 | 0.004 | -0.1477 | No | ||
| 51 | FZD1 | 16923 | 7933 | 0.004 | -0.1549 | No | ||
| 52 | WNT8B | 23841 | 8039 | 0.003 | -0.1605 | No | ||
| 53 | SOX17 | 14005 | 8214 | 0.003 | -0.1698 | No | ||
| 54 | CTBP1 | 16566 | 8222 | 0.003 | -0.1700 | No | ||
| 55 | FZD5 | 4743 | 8505 | 0.002 | -0.1852 | No | ||
| 56 | MYC | 22465 9435 | 9167 | 0.001 | -0.2209 | No | ||
| 57 | FZD2 | 12179 | 9189 | 0.001 | -0.2220 | No | ||
| 58 | WNT7A | 17068 | 9215 | 0.001 | -0.2233 | No | ||
| 59 | WNT4 | 16025 | 9307 | 0.001 | -0.2282 | No | ||
| 60 | PPARD | 9606 | 9325 | 0.001 | -0.2291 | No | ||
| 61 | FOSL1 | 23779 | 9674 | 0.000 | -0.2479 | No | ||
| 62 | SFRP5 | 23674 | 10001 | -0.000 | -0.2655 | No | ||
| 63 | FZD8 | 8990 | 10003 | -0.000 | -0.2656 | No | ||
| 64 | SFRP4 | 9806 | 10082 | -0.001 | -0.2698 | No | ||
| 65 | NFATC2 | 5168 2866 | 10189 | -0.001 | -0.2755 | No | ||
| 66 | PPP3CB | 5285 | 10274 | -0.001 | -0.2800 | No | ||
| 67 | WNT5A | 22066 5880 | 10412 | -0.001 | -0.2873 | No | ||
| 68 | CSNK2B | 23008 | 10540 | -0.001 | -0.2942 | No | ||
| 69 | NFATC3 | 5169 | 10711 | -0.002 | -0.3033 | No | ||
| 70 | WNT7B | 22172 | 10870 | -0.002 | -0.3118 | No | ||
| 71 | WNT3A | 20431 | 11322 | -0.003 | -0.3361 | No | ||
| 72 | FRAT2 | 23677 | 11545 | -0.003 | -0.3479 | No | ||
| 73 | NFATC4 | 22002 | 11897 | -0.004 | -0.3668 | No | ||
| 74 | PPP3R1 | 9611 489 | 12027 | -0.004 | -0.3736 | No | ||
| 75 | DKK2 | 15418 | 12032 | -0.004 | -0.3736 | No | ||
| 76 | FZD4 | 18193 | 12070 | -0.005 | -0.3755 | No | ||
| 77 | WNT10A | 14221 | 12124 | -0.005 | -0.3781 | No | ||
| 78 | PLCB1 | 14832 2821 | 12277 | -0.005 | -0.3862 | No | ||
| 79 | MMP7 | 19568 | 12321 | -0.005 | -0.3883 | No | ||
| 80 | SFRP2 | 15564 | 12389 | -0.005 | -0.3917 | No | ||
| 81 | WNT16 | 17512 1152 | 12569 | -0.006 | -0.4012 | No | ||
| 82 | WNT9A | 5709 | 12640 | -0.006 | -0.4048 | No | ||
| 83 | RAC3 | 20561 | 12968 | -0.007 | -0.4222 | No | ||
| 84 | APC2 | 10704 | 13014 | -0.007 | -0.4244 | No | ||
| 85 | FZD10 | 13565 16693 | 13131 | -0.008 | -0.4303 | No | ||
| 86 | WNT6 | 5881 | 13390 | -0.008 | -0.4440 | No | ||
| 87 | WNT1 | 22371 | 13491 | -0.009 | -0.4491 | No | ||
| 88 | CER1 | 15853 | 13876 | -0.011 | -0.4694 | No | ||
| 89 | DVL1 | 2412 15960 | 13914 | -0.011 | -0.4710 | No | ||
| 90 | DKK4 | 18653 | 13918 | -0.011 | -0.4708 | No | ||
| 91 | PRICKLE2 | 10837 | 13965 | -0.011 | -0.4729 | No | ||
| 92 | DAAM2 | 22945 176 | 14026 | -0.011 | -0.4757 | No | ||
| 93 | VANGL1 | 6034 | 14174 | -0.012 | -0.4832 | No | ||
| 94 | TCF7 | 1467 20466 | 14220 | -0.012 | -0.4851 | No | ||
| 95 | CCND1 | 4487 4488 8707 17535 | 14244 | -0.013 | -0.4859 | No | ||
| 96 | WNT8A | 1978 23603 | 14323 | -0.013 | -0.4897 | No | ||
| 97 | WIF1 | 19866 | 14396 | -0.014 | -0.4930 | No | ||
| 98 | FZD3 | 21784 | 14413 | -0.014 | -0.4934 | No | ||
| 99 | CAMK2G | 21905 | 14433 | -0.014 | -0.4939 | No | ||
| 100 | PORCN | 24190 | 14515 | -0.014 | -0.4977 | No | ||
| 101 | FRAT1 | 3681 23856 | 14875 | -0.018 | -0.5165 | No | ||
| 102 | PPP2CA | 20890 | 14993 | -0.019 | -0.5221 | No | ||
| 103 | GSK3B | 22761 | 15479 | -0.028 | -0.5473 | No | ||
| 104 | FZD7 | 14242 | 15480 | -0.028 | -0.5463 | No | ||
| 105 | PRICKLE1 | 8379 | 15596 | -0.031 | -0.5513 | No | ||
| 106 | DVL3 | 8873 22821 | 16146 | -0.053 | -0.5791 | No | ||
| 107 | ROCK1 | 5386 | 16614 | -0.085 | -0.6011 | No | ||
| 108 | PRKCA | 20174 | 16649 | -0.088 | -0.5997 | No | ||
| 109 | FZD6 | 22486 | 16681 | -0.091 | -0.5979 | No | ||
| 110 | MAPK9 | 1233 20903 1383 | 16779 | -0.101 | -0.5994 | No | ||
| 111 | CUL1 | 17462 | 16788 | -0.102 | -0.5960 | No | ||
| 112 | MAPK8 | 6459 | 16809 | -0.103 | -0.5932 | No | ||
| 113 | CSNK1E | 6570 2211 11332 | 16988 | -0.124 | -0.5982 | No | ||
| 114 | LRP6 | 9286 | 17152 | -0.146 | -0.6016 | No | ||
| 115 | PLCB3 | 23799 | 17365 | -0.181 | -0.6063 | Yes | ||
| 116 | ROCK2 | 21309 5387 | 17419 | -0.190 | -0.6021 | Yes | ||
| 117 | SENP2 | 7990 | 17626 | -0.231 | -0.6046 | Yes | ||
| 118 | CAMK2B | 20536 | 17631 | -0.233 | -0.5961 | Yes | ||
| 119 | CREBBP | 22682 8783 | 17830 | -0.281 | -0.5963 | Yes | ||
| 120 | LEF1 | 1860 15420 | 17935 | -0.318 | -0.5900 | Yes | ||
| 121 | PPP3CC | 21763 | 17966 | -0.329 | -0.5793 | Yes | ||
| 122 | AXIN1 | 1579 23330 | 17988 | -0.337 | -0.5678 | Yes | ||
| 123 | JUN | 15832 | 18245 | -0.472 | -0.5640 | Yes | ||
| 124 | CHD8 | 12581 | 18286 | -0.506 | -0.5473 | Yes | ||
| 125 | NLK | 5179 5178 | 18353 | -0.578 | -0.5292 | Yes | ||
| 126 | CCND2 | 16987 | 18450 | -0.739 | -0.5068 | Yes | ||
| 127 | CSNK2A2 | 4567 8808 | 18589 | -2.098 | -0.4358 | Yes | ||
| 128 | NFAT5 | 3921 7037 12036 | 18604 | -2.808 | -0.3315 | Yes | ||
| 129 | PRKX | 9619 5290 | 18605 | -2.856 | -0.2246 | Yes | ||
| 130 | RBX1 | 12130 7123 | 18609 | -2.999 | -0.1126 | Yes | ||
| 131 | TP53 | 20822 | 18610 | -3.019 | 0.0003 | Yes |