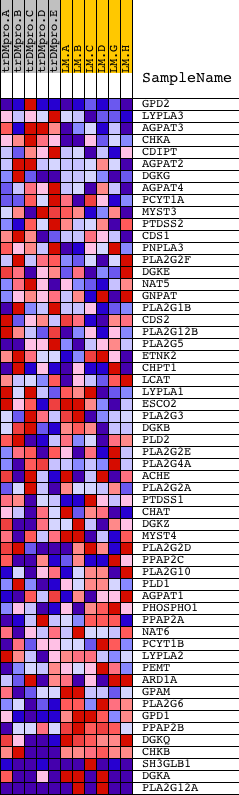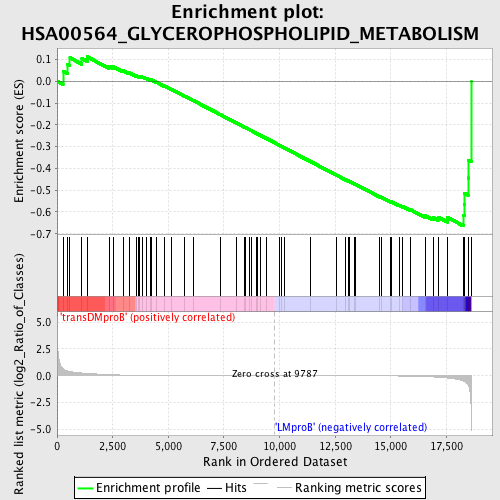
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA00564_GLYCEROPHOSPHOLIPID_METABOLISM |
| Enrichment Score (ES) | -0.6642164 |
| Normalized Enrichment Score (NES) | -1.418915 |
| Nominal p-value | 0.028103044 |
| FDR q-value | 0.7339541 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GPD2 | 4773 9014 | 266 | 0.627 | 0.0466 | No | ||
| 2 | LYPLA3 | 18482 | 479 | 0.426 | 0.0765 | No | ||
| 3 | AGPAT3 | 19713 | 578 | 0.378 | 0.1079 | No | ||
| 4 | CHKA | 23759 | 1115 | 0.256 | 0.1039 | No | ||
| 5 | CDIPT | 18071 | 1350 | 0.218 | 0.1125 | No | ||
| 6 | AGPAT2 | 2664 | 2334 | 0.101 | 0.0693 | No | ||
| 7 | DGKG | 4282 | 2545 | 0.086 | 0.0663 | No | ||
| 8 | AGPAT4 | 23384 | 2978 | 0.060 | 0.0488 | No | ||
| 9 | PCYT1A | 4572 | 3245 | 0.048 | 0.0391 | No | ||
| 10 | MYST3 | 18651 | 3567 | 0.038 | 0.0255 | No | ||
| 11 | PTDSS2 | 11334 | 3677 | 0.034 | 0.0229 | No | ||
| 12 | CDS1 | 16779 | 3710 | 0.034 | 0.0245 | No | ||
| 13 | PNPLA3 | 22406 | 3858 | 0.031 | 0.0195 | No | ||
| 14 | PLA2G2F | 15700 | 4012 | 0.027 | 0.0139 | No | ||
| 15 | DGKE | 7069 | 4206 | 0.024 | 0.0058 | No | ||
| 16 | NAT5 | 18601 7496 12597 | 4232 | 0.023 | 0.0067 | No | ||
| 17 | GNPAT | 18420 | 4460 | 0.020 | -0.0035 | No | ||
| 18 | PLA2G1B | 9581 | 4820 | 0.017 | -0.0212 | No | ||
| 19 | CDS2 | 14837 | 5160 | 0.014 | -0.0382 | No | ||
| 20 | PLA2G12B | 20016 | 5735 | 0.011 | -0.0680 | No | ||
| 21 | PLA2G5 | 9583 | 6122 | 0.009 | -0.0880 | No | ||
| 22 | ETNK2 | 5653 | 7353 | 0.005 | -0.1538 | No | ||
| 23 | CHPT1 | 3349 10015 | 8051 | 0.003 | -0.1910 | No | ||
| 24 | LCAT | 18762 | 8410 | 0.003 | -0.2100 | No | ||
| 25 | LYPLA1 | 4107 9580 | 8453 | 0.003 | -0.2120 | No | ||
| 26 | ESCO2 | 21781 | 8650 | 0.002 | -0.2224 | No | ||
| 27 | PLA2G3 | 20968 | 8718 | 0.002 | -0.2258 | No | ||
| 28 | DGKB | 5722 | 8953 | 0.002 | -0.2383 | No | ||
| 29 | PLD2 | 20803 | 9024 | 0.001 | -0.2419 | No | ||
| 30 | PLA2G2E | 16016 | 9132 | 0.001 | -0.2475 | No | ||
| 31 | PLA2G4A | 13809 | 9405 | 0.001 | -0.2621 | No | ||
| 32 | ACHE | 16665 | 9977 | -0.000 | -0.2928 | No | ||
| 33 | PLA2G2A | 16017 | 10085 | -0.001 | -0.2986 | No | ||
| 34 | PTDSS1 | 21616 | 10222 | -0.001 | -0.3058 | No | ||
| 35 | CHAT | 21882 | 11371 | -0.003 | -0.3674 | No | ||
| 36 | DGKZ | 2836 14522 | 12556 | -0.006 | -0.4306 | No | ||
| 37 | MYST4 | 7027 7026 12014 2739 | 12978 | -0.007 | -0.4526 | No | ||
| 38 | PLA2G2D | 16018 | 13076 | -0.007 | -0.4571 | No | ||
| 39 | PPAP2C | 6938 | 13153 | -0.008 | -0.4605 | No | ||
| 40 | PLA2G10 | 22658 | 13349 | -0.008 | -0.4702 | No | ||
| 41 | PLD1 | 15624 | 13419 | -0.009 | -0.4730 | No | ||
| 42 | AGPAT1 | 23275 | 14506 | -0.014 | -0.5302 | No | ||
| 43 | PHOSPHO1 | 10700 | 14578 | -0.015 | -0.5326 | No | ||
| 44 | PPAP2A | 21562 3258 3244 | 14990 | -0.019 | -0.5528 | No | ||
| 45 | NAT6 | 12131 | 15049 | -0.020 | -0.5540 | No | ||
| 46 | PCYT1B | 24289 | 15389 | -0.026 | -0.5697 | No | ||
| 47 | LYPLA2 | 15708 | 15536 | -0.029 | -0.5748 | No | ||
| 48 | PEMT | 20423 | 15897 | -0.041 | -0.5902 | No | ||
| 49 | ARD1A | 2555 7089 12092 | 16557 | -0.080 | -0.6179 | No | ||
| 50 | GPAM | 4795 | 16906 | -0.114 | -0.6256 | Yes | ||
| 51 | PLA2G6 | 22209 | 17123 | -0.143 | -0.6234 | Yes | ||
| 52 | GPD1 | 22361 | 17548 | -0.217 | -0.6252 | Yes | ||
| 53 | PPAP2B | 7503 16161 | 18273 | -0.497 | -0.6160 | Yes | ||
| 54 | DGKQ | 4287 | 18307 | -0.526 | -0.5666 | Yes | ||
| 55 | CHKB | 4518 | 18321 | -0.547 | -0.5142 | Yes | ||
| 56 | SH3GLB1 | 12057 7051 | 18473 | -0.809 | -0.4438 | Yes | ||
| 57 | DGKA | 3359 19589 | 18482 | -0.843 | -0.3623 | Yes | ||
| 58 | PLA2G12A | 1782 7281 1839 | 18613 | -3.804 | 0.0002 | Yes |