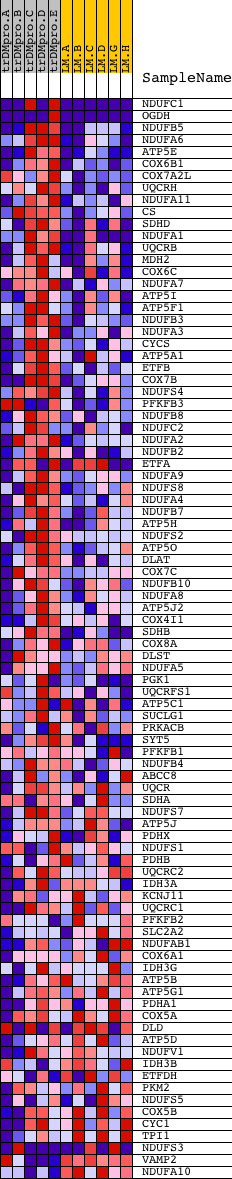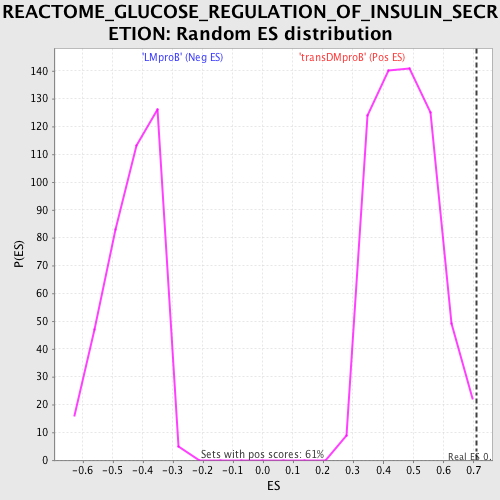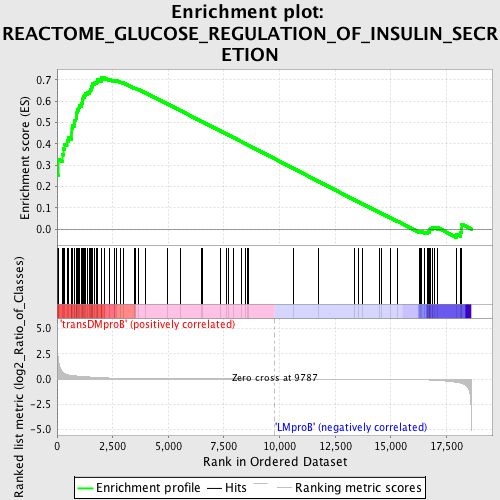
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | REACTOME_GLUCOSE_REGULATION_OF_INSULIN_SECRETION |
| Enrichment Score (ES) | 0.71075314 |
| Normalized Enrichment Score (NES) | 1.5034847 |
| Nominal p-value | 0.0016393443 |
| FDR q-value | 0.57327676 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NDUFC1 | 7287 12339 | 0 | 5.436 | 0.2552 | Yes | ||
| 2 | OGDH | 9502 1412 | 76 | 1.582 | 0.3254 | Yes | ||
| 3 | NDUFB5 | 12277 | 229 | 0.698 | 0.3500 | Yes | ||
| 4 | NDUFA6 | 12473 | 275 | 0.620 | 0.3766 | Yes | ||
| 5 | ATP5E | 14321 | 326 | 0.541 | 0.3993 | Yes | ||
| 6 | COX6B1 | 4283 8479 | 445 | 0.445 | 0.4138 | Yes | ||
| 7 | COX7A2L | 9819 | 525 | 0.401 | 0.4284 | Yes | ||
| 8 | UQCRH | 12378 | 654 | 0.356 | 0.4382 | Yes | ||
| 9 | NDUFA11 | 12832 | 657 | 0.354 | 0.4547 | Yes | ||
| 10 | CS | 19839 | 668 | 0.349 | 0.4706 | Yes | ||
| 11 | SDHD | 19125 | 687 | 0.345 | 0.4858 | Yes | ||
| 12 | NDUFA1 | 24170 | 765 | 0.323 | 0.4968 | Yes | ||
| 13 | UQCRB | 12545 | 779 | 0.320 | 0.5111 | Yes | ||
| 14 | MDH2 | 9399 | 849 | 0.305 | 0.5217 | Yes | ||
| 15 | COX6C | 8775 | 873 | 0.300 | 0.5345 | Yes | ||
| 16 | NDUFA7 | 12347 | 888 | 0.298 | 0.5477 | Yes | ||
| 17 | ATP5I | 8637 | 905 | 0.294 | 0.5607 | Yes | ||
| 18 | ATP5F1 | 15212 | 982 | 0.282 | 0.5698 | Yes | ||
| 19 | NDUFB3 | 12362 | 1009 | 0.275 | 0.5813 | Yes | ||
| 20 | NDUFA3 | 18410 | 1107 | 0.257 | 0.5882 | Yes | ||
| 21 | CYCS | 8821 | 1127 | 0.254 | 0.5990 | Yes | ||
| 22 | ATP5A1 | 23505 | 1134 | 0.252 | 0.6105 | Yes | ||
| 23 | ETFB | 18279 | 1164 | 0.248 | 0.6206 | Yes | ||
| 24 | COX7B | 7260 | 1233 | 0.233 | 0.6279 | Yes | ||
| 25 | NDUFS4 | 9452 5157 3173 | 1272 | 0.228 | 0.6366 | Yes | ||
| 26 | PFKFB3 | 2748 | 1365 | 0.216 | 0.6418 | Yes | ||
| 27 | NDUFB8 | 7418 | 1441 | 0.205 | 0.6473 | Yes | ||
| 28 | NDUFC2 | 18183 | 1482 | 0.199 | 0.6545 | Yes | ||
| 29 | NDUFA2 | 9450 | 1526 | 0.192 | 0.6612 | Yes | ||
| 30 | NDUFB2 | 7542 | 1536 | 0.191 | 0.6697 | Yes | ||
| 31 | ETFA | 4292 8495 3155 | 1598 | 0.181 | 0.6749 | Yes | ||
| 32 | NDUFA9 | 12288 | 1608 | 0.179 | 0.6828 | Yes | ||
| 33 | NDUFS8 | 23950 | 1700 | 0.169 | 0.6858 | Yes | ||
| 34 | NDUFA4 | 9451 | 1786 | 0.159 | 0.6886 | Yes | ||
| 35 | NDUFB7 | 12429 | 1797 | 0.158 | 0.6955 | Yes | ||
| 36 | ATP5H | 12948 | 1804 | 0.157 | 0.7025 | Yes | ||
| 37 | NDUFS2 | 4089 13760 | 1978 | 0.135 | 0.6995 | Yes | ||
| 38 | ATP5O | 22539 | 1996 | 0.133 | 0.7049 | Yes | ||
| 39 | DLAT | 19123 | 2003 | 0.132 | 0.7108 | Yes | ||
| 40 | COX7C | 8776 | 2137 | 0.118 | 0.7091 | No | ||
| 41 | NDUFB10 | 23084 | 2337 | 0.100 | 0.7031 | No | ||
| 42 | NDUFA8 | 14605 | 2559 | 0.085 | 0.6951 | No | ||
| 43 | ATP5J2 | 12186 | 2580 | 0.084 | 0.6980 | No | ||
| 44 | COX4I1 | 18444 | 2649 | 0.079 | 0.6980 | No | ||
| 45 | SDHB | 2348 12566 14880 | 2862 | 0.065 | 0.6896 | No | ||
| 46 | COX8A | 8777 | 2965 | 0.060 | 0.6869 | No | ||
| 47 | DLST | 8131 | 3479 | 0.040 | 0.6611 | No | ||
| 48 | NDUFA5 | 1076 7544 | 3509 | 0.039 | 0.6614 | No | ||
| 49 | PGK1 | 9557 5244 | 3674 | 0.034 | 0.6541 | No | ||
| 50 | UQCRFS1 | 21499 | 3972 | 0.028 | 0.6394 | No | ||
| 51 | ATP5C1 | 8635 | 4941 | 0.016 | 0.5879 | No | ||
| 52 | SUCLG1 | 17409 1059 | 5527 | 0.012 | 0.5569 | No | ||
| 53 | PRKACB | 15140 | 6469 | 0.008 | 0.5065 | No | ||
| 54 | SYT5 | 17986 | 6539 | 0.007 | 0.5031 | No | ||
| 55 | PFKFB1 | 9552 | 7348 | 0.005 | 0.4597 | No | ||
| 56 | NDUFB4 | 22596 7541 | 7629 | 0.004 | 0.4448 | No | ||
| 57 | ABCC8 | 9941 11649 | 7713 | 0.004 | 0.4405 | No | ||
| 58 | UQCR | 12382 | 7936 | 0.004 | 0.4287 | No | ||
| 59 | SDHA | 12436 | 8271 | 0.003 | 0.4108 | No | ||
| 60 | NDUFS7 | 19946 | 8476 | 0.002 | 0.3999 | No | ||
| 61 | ATP5J | 880 22555 | 8543 | 0.002 | 0.3965 | No | ||
| 62 | PDHX | 14510 | 8609 | 0.002 | 0.3931 | No | ||
| 63 | NDUFS1 | 13940 | 10638 | -0.002 | 0.2837 | No | ||
| 64 | PDHB | 12670 7548 | 11756 | -0.004 | 0.2236 | No | ||
| 65 | UQCRC2 | 18102 | 13373 | -0.008 | 0.1367 | No | ||
| 66 | IDH3A | 19447 | 13566 | -0.009 | 0.1268 | No | ||
| 67 | KCNJ11 | 17822 | 13725 | -0.010 | 0.1187 | No | ||
| 68 | UQCRC1 | 10259 | 14484 | -0.014 | 0.0785 | No | ||
| 69 | PFKFB2 | 4116 5242 | 14566 | -0.015 | 0.0748 | No | ||
| 70 | SLC2A2 | 15623 1880 1855 | 14974 | -0.019 | 0.0537 | No | ||
| 71 | NDUFAB1 | 7667 | 15305 | -0.024 | 0.0371 | No | ||
| 72 | COX6A1 | 8774 4553 | 16268 | -0.060 | -0.0121 | No | ||
| 73 | IDH3G | 24132 | 16333 | -0.064 | -0.0125 | No | ||
| 74 | ATP5B | 19846 | 16374 | -0.067 | -0.0115 | No | ||
| 75 | ATP5G1 | 8636 | 16531 | -0.077 | -0.0163 | No | ||
| 76 | PDHA1 | 24020 | 16635 | -0.087 | -0.0178 | No | ||
| 77 | COX5A | 19431 | 16647 | -0.088 | -0.0142 | No | ||
| 78 | DLD | 2097 21090 | 16671 | -0.090 | -0.0112 | No | ||
| 79 | ATP5D | 19949 | 16740 | -0.097 | -0.0104 | No | ||
| 80 | NDUFV1 | 23953 | 16742 | -0.097 | -0.0059 | No | ||
| 81 | IDH3B | 9299 | 16756 | -0.099 | -0.0019 | No | ||
| 82 | ETFDH | 15313 | 16787 | -0.102 | 0.0012 | No | ||
| 83 | PKM2 | 3642 9573 | 16872 | -0.110 | 0.0018 | No | ||
| 84 | NDUFS5 | 9296 | 16878 | -0.110 | 0.0067 | No | ||
| 85 | COX5B | 4552 | 16962 | -0.121 | 0.0079 | No | ||
| 86 | CYC1 | 12348 | 17113 | -0.141 | 0.0064 | No | ||
| 87 | TPI1 | 5795 10212 | 17955 | -0.325 | -0.0237 | No | ||
| 88 | NDUFS3 | 7553 12683 | 18145 | -0.404 | -0.0150 | No | ||
| 89 | VAMP2 | 20826 | 18175 | -0.423 | 0.0033 | No | ||
| 90 | NDUFA10 | 7420 | 18192 | -0.435 | 0.0229 | No |