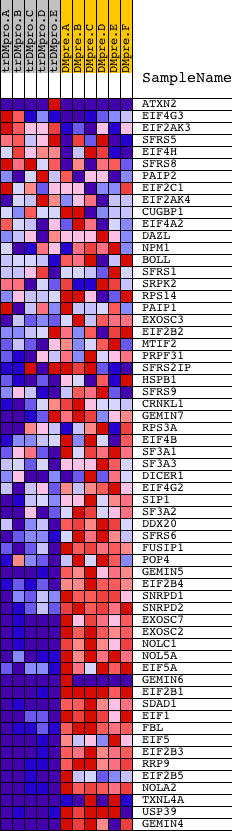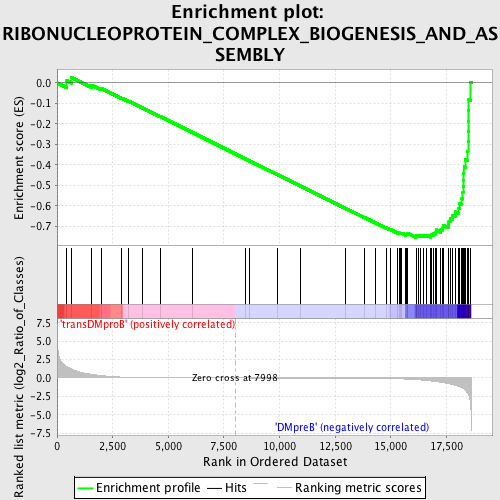
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | RIBONUCLEOPROTEIN_COMPLEX_BIOGENESIS_AND_ASSEMBLY |
| Enrichment Score (ES) | -0.7538345 |
| Normalized Enrichment Score (NES) | -1.6606301 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.04350018 |
| FWER p-Value | 0.163 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATXN2 | 9780 3506 | 440 | 1.551 | 0.0107 | No | ||
| 2 | EIF4G3 | 10517 | 646 | 1.196 | 0.0263 | No | ||
| 3 | EIF2AK3 | 17421 | 1545 | 0.505 | -0.0109 | No | ||
| 4 | SFRS5 | 9808 2062 | 1972 | 0.334 | -0.0265 | No | ||
| 5 | EIF4H | 16352 | 2905 | 0.118 | -0.0741 | No | ||
| 6 | SFRS8 | 10543 6089 | 3220 | 0.082 | -0.0892 | No | ||
| 7 | PAIP2 | 12593 | 3816 | 0.041 | -0.1203 | No | ||
| 8 | EIF2C1 | 10672 | 4645 | 0.016 | -0.1646 | No | ||
| 9 | EIF2AK4 | 14909 | 6099 | 0.005 | -0.2428 | No | ||
| 10 | CUGBP1 | 2805 8819 4576 2924 | 8484 | -0.001 | -0.3712 | No | ||
| 11 | EIF4A2 | 4660 1679 1645 | 8669 | -0.002 | -0.3811 | No | ||
| 12 | DAZL | 22944 | 9899 | -0.005 | -0.4472 | No | ||
| 13 | NPM1 | 1196 | 10938 | -0.008 | -0.5030 | No | ||
| 14 | BOLL | 7960 | 12946 | -0.020 | -0.6107 | No | ||
| 15 | SFRS1 | 8492 | 13823 | -0.033 | -0.6572 | No | ||
| 16 | SRPK2 | 5513 | 14328 | -0.045 | -0.6833 | No | ||
| 17 | RPS14 | 9751 | 14793 | -0.063 | -0.7069 | No | ||
| 18 | PAIP1 | 21556 | 14971 | -0.073 | -0.7148 | No | ||
| 19 | EXOSC3 | 15890 | 15302 | -0.098 | -0.7304 | No | ||
| 20 | EIF2B2 | 21204 | 15373 | -0.106 | -0.7319 | No | ||
| 21 | MTIF2 | 13354 | 15453 | -0.115 | -0.7336 | No | ||
| 22 | PRPF31 | 7594 | 15484 | -0.118 | -0.7326 | No | ||
| 23 | SFRS2IP | 7794 13009 | 15677 | -0.143 | -0.7397 | No | ||
| 24 | HSPB1 | 4879 | 15693 | -0.146 | -0.7373 | No | ||
| 25 | SFRS9 | 16731 | 15758 | -0.155 | -0.7373 | No | ||
| 26 | CRNKL1 | 14407 | 15765 | -0.156 | -0.7342 | No | ||
| 27 | GEMIN7 | 17944 | 16131 | -0.222 | -0.7489 | Yes | ||
| 28 | RPS3A | 9755 | 16163 | -0.228 | -0.7455 | Yes | ||
| 29 | EIF4B | 13279 7979 | 16251 | -0.250 | -0.7446 | Yes | ||
| 30 | SF3A1 | 7450 | 16323 | -0.267 | -0.7425 | Yes | ||
| 31 | SF3A3 | 16091 | 16480 | -0.305 | -0.7442 | Yes | ||
| 32 | DICER1 | 20989 | 16607 | -0.341 | -0.7434 | Yes | ||
| 33 | EIF4G2 | 1908 8892 | 16768 | -0.389 | -0.7434 | Yes | ||
| 34 | SIP1 | 21263 | 16833 | -0.408 | -0.7378 | Yes | ||
| 35 | SF3A2 | 19938 | 16915 | -0.442 | -0.7323 | Yes | ||
| 36 | DDX20 | 15213 | 17024 | -0.479 | -0.7275 | Yes | ||
| 37 | SFRS6 | 14751 | 17031 | -0.480 | -0.7172 | Yes | ||
| 38 | FUSIP1 | 4715 16036 | 17251 | -0.577 | -0.7161 | Yes | ||
| 39 | POP4 | 7261 | 17344 | -0.618 | -0.7074 | Yes | ||
| 40 | GEMIN5 | 20439 | 17347 | -0.619 | -0.6937 | Yes | ||
| 41 | EIF2B4 | 16574 | 17589 | -0.755 | -0.6899 | Yes | ||
| 42 | SNRPD1 | 23622 | 17611 | -0.767 | -0.6740 | Yes | ||
| 43 | SNRPD2 | 8412 | 17693 | -0.836 | -0.6598 | Yes | ||
| 44 | EXOSC7 | 19256 | 17784 | -0.900 | -0.6447 | Yes | ||
| 45 | EXOSC2 | 15044 | 17897 | -0.967 | -0.6292 | Yes | ||
| 46 | NOLC1 | 7704 | 18044 | -1.129 | -0.6120 | Yes | ||
| 47 | NOL5A | 12474 | 18075 | -1.164 | -0.5877 | Yes | ||
| 48 | EIF5A | 11345 20379 6590 | 18187 | -1.340 | -0.5639 | Yes | ||
| 49 | GEMIN6 | 12492 7415 | 18232 | -1.391 | -0.5354 | Yes | ||
| 50 | EIF2B1 | 16368 3458 | 18250 | -1.416 | -0.5049 | Yes | ||
| 51 | SDAD1 | 16479 | 18273 | -1.454 | -0.4737 | Yes | ||
| 52 | EIF1 | 23563 | 18278 | -1.468 | -0.4413 | Yes | ||
| 53 | FBL | 8955 | 18289 | -1.496 | -0.4086 | Yes | ||
| 54 | EIF5 | 5736 | 18356 | -1.677 | -0.3749 | Yes | ||
| 55 | EIF2B3 | 16118 | 18455 | -2.083 | -0.3339 | Yes | ||
| 56 | RRP9 | 19328 | 18469 | -2.146 | -0.2870 | Yes | ||
| 57 | EIF2B5 | 1719 22822 | 18488 | -2.245 | -0.2380 | Yes | ||
| 58 | NOLA2 | 11972 | 18500 | -2.357 | -0.1863 | Yes | ||
| 59 | TXNL4A | 6567 11329 | 18503 | -2.361 | -0.1339 | Yes | ||
| 60 | USP39 | 1116 1083 11373 | 18509 | -2.419 | -0.0804 | Yes | ||
| 61 | GEMIN4 | 6592 6591 | 18592 | -3.878 | 0.0013 | Yes |