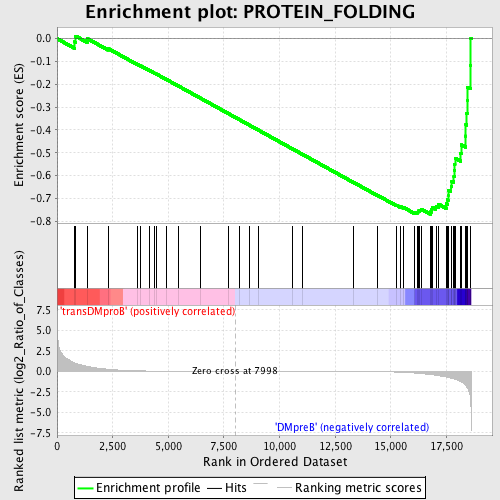
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | PROTEIN_FOLDING |
| Enrichment Score (ES) | -0.7702276 |
| Normalized Enrichment Score (NES) | -1.6479328 |
| Nominal p-value | 0.0045045046 |
| FDR q-value | 0.042078458 |
| FWER p-Value | 0.225 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AIP | 23955 | 796 | 1.002 | -0.0141 | No | ||
| 2 | LRPAP1 | 5005 | 837 | 0.961 | 0.0113 | No | ||
| 3 | TBCC | 23215 | 1352 | 0.597 | 0.0007 | No | ||
| 4 | TBCA | 10037 | 2289 | 0.245 | -0.0427 | No | ||
| 5 | PDIA6 | 21308 | 3623 | 0.051 | -0.1130 | No | ||
| 6 | HSPA4L | 5214 | 3754 | 0.044 | -0.1188 | No | ||
| 7 | FKBP5 | 23051 | 4165 | 0.027 | -0.1401 | No | ||
| 8 | PPIA | 1284 11188 | 4398 | 0.021 | -0.1520 | No | ||
| 9 | CCT6B | 20326 | 4474 | 0.020 | -0.1554 | No | ||
| 10 | PDIA2 | 23062 22604 1702 | 4925 | 0.013 | -0.1793 | No | ||
| 11 | DNAJA1 | 4878 16242 2345 | 5454 | 0.008 | -0.2075 | No | ||
| 12 | DNAJB6 | 6246 3538 | 6463 | 0.004 | -0.2617 | No | ||
| 13 | MKKS | 2946 14416 | 7719 | 0.001 | -0.3293 | No | ||
| 14 | APCS | 13748 | 8184 | -0.000 | -0.3543 | No | ||
| 15 | FKBP9 | 17433 | 8648 | -0.002 | -0.3791 | No | ||
| 16 | NFYC | 9460 | 9047 | -0.003 | -0.4005 | No | ||
| 17 | FKBP6 | 16344 | 10579 | -0.007 | -0.4828 | No | ||
| 18 | TBCE | 21543 | 11032 | -0.008 | -0.5069 | No | ||
| 19 | ERO1L | 6930 11924 6929 | 13311 | -0.025 | -0.6289 | No | ||
| 20 | PIGK | 1815 6834 11629 | 14411 | -0.047 | -0.6867 | No | ||
| 21 | APOA2 | 8615 4044 | 15270 | -0.095 | -0.7302 | No | ||
| 22 | STUB1 | 23070 | 15416 | -0.112 | -0.7348 | No | ||
| 23 | UGCGL1 | 6721 | 15555 | -0.127 | -0.7386 | No | ||
| 24 | PIN4 | 12817 | 16064 | -0.210 | -0.7600 | Yes | ||
| 25 | CLPX | 19403 | 16183 | -0.233 | -0.7596 | Yes | ||
| 26 | TXNDC4 | 15886 | 16223 | -0.243 | -0.7548 | Yes | ||
| 27 | LMAN1 | 12867 | 16303 | -0.262 | -0.7515 | Yes | ||
| 28 | CLN3 | 17635 | 16364 | -0.275 | -0.7469 | Yes | ||
| 29 | TOR1A | 11395 2909 | 16799 | -0.397 | -0.7589 | Yes | ||
| 30 | TBCD | 20555 | 16816 | -0.403 | -0.7482 | Yes | ||
| 31 | CCT4 | 8710 | 16853 | -0.415 | -0.7382 | Yes | ||
| 32 | CCT7 | 17385 | 17030 | -0.479 | -0.7339 | Yes | ||
| 33 | ARL2 | 23992 | 17151 | -0.528 | -0.7252 | Yes | ||
| 34 | DNAJC7 | 20227 | 17490 | -0.691 | -0.7236 | Yes | ||
| 35 | TTC1 | 7342 | 17548 | -0.730 | -0.7058 | Yes | ||
| 36 | BAG4 | 7431 | 17577 | -0.748 | -0.6858 | Yes | ||
| 37 | ST13 | 12865 | 17583 | -0.750 | -0.6646 | Yes | ||
| 38 | FKBP4 | 4726 | 17704 | -0.838 | -0.6470 | Yes | ||
| 39 | AHSA1 | 21192 | 17740 | -0.864 | -0.6241 | Yes | ||
| 40 | PPIH | 12287 | 17835 | -0.930 | -0.6025 | Yes | ||
| 41 | GLRX2 | 12795 | 17859 | -0.940 | -0.5767 | Yes | ||
| 42 | ERP29 | 12524 | 17871 | -0.946 | -0.5502 | Yes | ||
| 43 | PFDN4 | 4260 | 17885 | -0.957 | -0.5234 | Yes | ||
| 44 | RUVBL2 | 9766 | 18134 | -1.250 | -0.5009 | Yes | ||
| 45 | CCT6A | 16689 | 18180 | -1.327 | -0.4653 | Yes | ||
| 46 | HSPE1 | 9133 | 18364 | -1.702 | -0.4263 | Yes | ||
| 47 | CCT3 | 8709 | 18372 | -1.727 | -0.3771 | Yes | ||
| 48 | SEP15 | 13527 1895 15398 | 18404 | -1.823 | -0.3265 | Yes | ||
| 49 | BAG5 | 20981 192 | 18434 | -2.008 | -0.2705 | Yes | ||
| 50 | BAG2 | 13985 | 18442 | -2.037 | -0.2124 | Yes | ||
| 51 | DNAJA3 | 1732 13518 | 18586 | -3.642 | -0.1157 | Yes | ||
| 52 | HSP90AA1 | 4883 4882 9131 | 18597 | -4.087 | 0.0010 | Yes |

