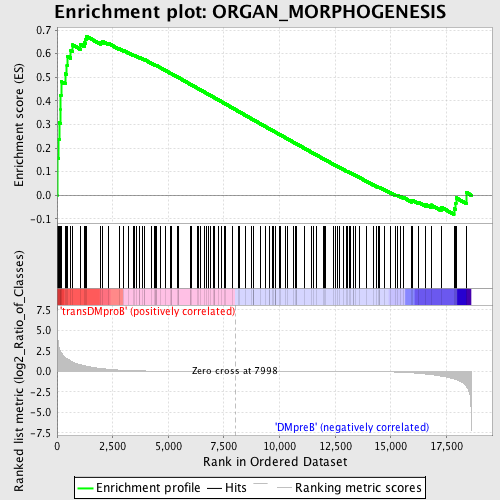
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | ORGAN_MORPHOGENESIS |
| Enrichment Score (ES) | 0.6742078 |
| Normalized Enrichment Score (NES) | 1.5749395 |
| Nominal p-value | 0.0017452007 |
| FDR q-value | 0.19654687 |
| FWER p-Value | 0.684 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MYH9 | 2252 2244 | 4 | 6.155 | 0.1595 | Yes | ||
| 2 | SERPINF1 | 9793 | 69 | 3.106 | 0.2367 | Yes | ||
| 3 | THY1 | 19481 | 93 | 2.773 | 0.3074 | Yes | ||
| 4 | ENG | 4667 | 160 | 2.332 | 0.3644 | Yes | ||
| 5 | DGCR2 | 4628 8851 | 164 | 2.319 | 0.4244 | Yes | ||
| 6 | RHOB | 4411 | 181 | 2.245 | 0.4818 | Yes | ||
| 7 | VEGFA | 22969 | 368 | 1.660 | 0.5149 | Yes | ||
| 8 | CUL7 | 23217 | 442 | 1.547 | 0.5511 | Yes | ||
| 9 | TGFB1 | 18332 | 479 | 1.483 | 0.5876 | Yes | ||
| 10 | EGFL7 | 87 11687 2706 | 615 | 1.245 | 0.6126 | Yes | ||
| 11 | RASA1 | 10174 | 698 | 1.106 | 0.6369 | Yes | ||
| 12 | STX2 | 3583 16365 | 1059 | 0.793 | 0.6380 | Yes | ||
| 13 | RUNX1 | 4481 | 1221 | 0.667 | 0.6466 | Yes | ||
| 14 | EMCN | 7211 | 1286 | 0.627 | 0.6594 | Yes | ||
| 15 | ATPIF1 | 4429 | 1310 | 0.617 | 0.6742 | Yes | ||
| 16 | SOD1 | 9846 | 1945 | 0.341 | 0.6488 | No | ||
| 17 | CCM2 | 20950 | 2040 | 0.312 | 0.6518 | No | ||
| 18 | EGF | 15169 | 2310 | 0.241 | 0.6435 | No | ||
| 19 | FGD1 | 24228 | 2792 | 0.137 | 0.6211 | No | ||
| 20 | FLI1 | 4729 | 2988 | 0.107 | 0.6133 | No | ||
| 21 | NF1 | 5165 | 3219 | 0.082 | 0.6030 | No | ||
| 22 | IL7 | 4921 | 3442 | 0.063 | 0.5926 | No | ||
| 23 | TLE1 | 5765 10185 15859 | 3478 | 0.061 | 0.5923 | No | ||
| 24 | PAX5 | 15892 | 3564 | 0.055 | 0.5891 | No | ||
| 25 | TLE3 | 5767 19416 | 3711 | 0.046 | 0.5824 | No | ||
| 26 | PF4 | 16800 | 3714 | 0.046 | 0.5835 | No | ||
| 27 | MYH7 | 21828 4709 | 3836 | 0.040 | 0.5780 | No | ||
| 28 | C1GALT1 | 8246 | 3933 | 0.036 | 0.5737 | No | ||
| 29 | PKD2 | 16772 | 3944 | 0.035 | 0.5741 | No | ||
| 30 | VANGL2 | 4109 8220 | 4255 | 0.025 | 0.5580 | No | ||
| 31 | IL18 | 9172 | 4373 | 0.022 | 0.5522 | No | ||
| 32 | ABLIM1 | 3717 3766 3679 | 4429 | 0.021 | 0.5498 | No | ||
| 33 | ROBO4 | 7895 3064 | 4440 | 0.020 | 0.5498 | No | ||
| 34 | GYPC | 23474 | 4445 | 0.020 | 0.5501 | No | ||
| 35 | HCCS | 9079 4842 | 4459 | 0.020 | 0.5499 | No | ||
| 36 | PROK2 | 17057 | 4625 | 0.017 | 0.5414 | No | ||
| 37 | CCL2 | 9788 | 4864 | 0.013 | 0.5289 | No | ||
| 38 | PITX2 | 15424 1878 | 5080 | 0.011 | 0.5176 | No | ||
| 39 | STAB1 | 21892 | 5143 | 0.011 | 0.5145 | No | ||
| 40 | ACVRL1 | 22354 | 5159 | 0.011 | 0.5140 | No | ||
| 41 | PLAC1 | 12079 | 5400 | 0.009 | 0.5012 | No | ||
| 42 | COL4A2 | 18688 | 5467 | 0.008 | 0.4978 | No | ||
| 43 | ANGPTL3 | 16171 | 5473 | 0.008 | 0.4978 | No | ||
| 44 | PITX3 | 9571 | 6008 | 0.006 | 0.4691 | No | ||
| 45 | RORB | 10356 | 6016 | 0.006 | 0.4688 | No | ||
| 46 | EPGN | 16798 | 6039 | 0.006 | 0.4678 | No | ||
| 47 | FOXN1 | 20337 | 6317 | 0.005 | 0.4529 | No | ||
| 48 | TINAG | 3141 19060 | 6337 | 0.004 | 0.4520 | No | ||
| 49 | NPPB | 15994 | 6431 | 0.004 | 0.4471 | No | ||
| 50 | ITGAX | 18058 | 6630 | 0.003 | 0.4365 | No | ||
| 51 | FGF16 | 24270 | 6721 | 0.003 | 0.4317 | No | ||
| 52 | SFTPB | 17414 | 6818 | 0.003 | 0.4266 | No | ||
| 53 | COMP | 18589 | 6879 | 0.003 | 0.4234 | No | ||
| 54 | PAX3 | 13910 | 7016 | 0.002 | 0.4161 | No | ||
| 55 | EREG | 4679 16797 | 7087 | 0.002 | 0.4124 | No | ||
| 56 | SIX6 | 21247 | 7094 | 0.002 | 0.4121 | No | ||
| 57 | SPRY2 | 21725 | 7275 | 0.002 | 0.4024 | No | ||
| 58 | HEY2 | 19794 | 7399 | 0.001 | 0.3958 | No | ||
| 59 | NOTCH4 | 23277 9475 | 7543 | 0.001 | 0.3881 | No | ||
| 60 | APLP1 | 17890 | 7566 | 0.001 | 0.3869 | No | ||
| 61 | NKX6-1 | 16463 | 7883 | 0.000 | 0.3698 | No | ||
| 62 | LHX1 | 4995 | 8140 | -0.000 | 0.3560 | No | ||
| 63 | SHH | 16581 | 8203 | -0.001 | 0.3527 | No | ||
| 64 | LSR | 17877 2176 | 8459 | -0.001 | 0.3389 | No | ||
| 65 | EPHB4 | 4676 | 8759 | -0.002 | 0.3228 | No | ||
| 66 | TBX1 | 10039 5630 | 8806 | -0.002 | 0.3203 | No | ||
| 67 | PTCH1 | 5304 | 8817 | -0.002 | 0.3198 | No | ||
| 68 | FABP1 | 17420 | 8820 | -0.002 | 0.3198 | No | ||
| 69 | IL17F | 13992 | 9150 | -0.003 | 0.3021 | No | ||
| 70 | AMOT | 11341 24037 6587 | 9351 | -0.003 | 0.2913 | No | ||
| 71 | ONECUT2 | 10346 | 9540 | -0.004 | 0.2813 | No | ||
| 72 | PDX1 | 16621 | 9556 | -0.004 | 0.2805 | No | ||
| 73 | MSX1 | 16547 | 9674 | -0.004 | 0.2743 | No | ||
| 74 | GLMN | 16452 3562 | 9712 | -0.004 | 0.2724 | No | ||
| 75 | PLG | 1593 23383 | 9797 | -0.004 | 0.2680 | No | ||
| 76 | SCG2 | 5410 14425 | 9812 | -0.004 | 0.2674 | No | ||
| 77 | NPR1 | 9480 | 9833 | -0.005 | 0.2664 | No | ||
| 78 | COL9A1 | 3950 4549 | 9996 | -0.005 | 0.2578 | No | ||
| 79 | PAX6 | 5223 | 10062 | -0.005 | 0.2544 | No | ||
| 80 | FOXC1 | 21676 | 10271 | -0.006 | 0.2433 | No | ||
| 81 | COL4A3 | 8766 | 10340 | -0.006 | 0.2398 | No | ||
| 82 | ZIC1 | 19040 | 10643 | -0.007 | 0.2236 | No | ||
| 83 | TNFSF12 | 10209 | 10724 | -0.007 | 0.2195 | No | ||
| 84 | FGF10 | 4719 8962 | 10747 | -0.007 | 0.2185 | No | ||
| 85 | DCN | 19897 4601 8840 3366 | 10767 | -0.007 | 0.2176 | No | ||
| 86 | CRX | 17961 4694 | 11115 | -0.009 | 0.1991 | No | ||
| 87 | SPHK1 | 5484 1266 1402 | 11415 | -0.010 | 0.1832 | No | ||
| 88 | DSCAML1 | 4338 13224 | 11504 | -0.010 | 0.1787 | No | ||
| 89 | COL18A1 | 19719 3360 | 11656 | -0.011 | 0.1708 | No | ||
| 90 | CHRNA7 | 17808 | 11963 | -0.012 | 0.1546 | No | ||
| 91 | TLE2 | 3378 | 12031 | -0.013 | 0.1513 | No | ||
| 92 | POU1F1 | 5257 | 12059 | -0.013 | 0.1501 | No | ||
| 93 | SPINK5 | 23569 | 12405 | -0.016 | 0.1319 | No | ||
| 94 | NOTCH2 | 15485 | 12491 | -0.016 | 0.1277 | No | ||
| 95 | LHX3 | 2761 4996 | 12605 | -0.017 | 0.1220 | No | ||
| 96 | MYH6 | 9436 | 12697 | -0.018 | 0.1176 | No | ||
| 97 | PAX4 | 17199 | 12891 | -0.020 | 0.1077 | No | ||
| 98 | FGF2 | 15608 | 13003 | -0.021 | 0.1022 | No | ||
| 99 | LY6H | 6244 22256 10732 | 13070 | -0.022 | 0.0992 | No | ||
| 100 | ELN | 8896 | 13122 | -0.022 | 0.0970 | No | ||
| 101 | TGFB3 | 10161 | 13172 | -0.023 | 0.0950 | No | ||
| 102 | GAA | 20575 | 13324 | -0.025 | 0.0874 | No | ||
| 103 | COL5A3 | 19219 | 13332 | -0.025 | 0.0877 | No | ||
| 104 | AES | 19925 478 | 13431 | -0.026 | 0.0831 | No | ||
| 105 | CDX1 | 23429 | 13584 | -0.029 | 0.0756 | No | ||
| 106 | PML | 3015 3074 3020 5270 | 13898 | -0.034 | 0.0596 | No | ||
| 107 | TBX3 | 16723 | 14201 | -0.041 | 0.0443 | No | ||
| 108 | CDH13 | 4506 3826 | 14375 | -0.046 | 0.0361 | No | ||
| 109 | TBX4 | 5633 | 14442 | -0.048 | 0.0338 | No | ||
| 110 | GLI2 | 13859 | 14477 | -0.049 | 0.0333 | No | ||
| 111 | HEY1 | 15378 | 14499 | -0.051 | 0.0334 | No | ||
| 112 | CANX | 4474 | 14714 | -0.060 | 0.0234 | No | ||
| 113 | HTATIP2 | 18232 | 14987 | -0.074 | 0.0106 | No | ||
| 114 | MYBPC3 | 9434 | 15228 | -0.092 | 0.0000 | No | ||
| 115 | NCL | 5153 13899 | 15284 | -0.096 | -0.0004 | No | ||
| 116 | SYK | 21636 | 15422 | -0.112 | -0.0049 | No | ||
| 117 | PCSK9 | 15822 | 15587 | -0.131 | -0.0104 | No | ||
| 118 | PROX1 | 9623 | 15932 | -0.187 | -0.0242 | No | ||
| 119 | DGCR6 | 22831 | 15992 | -0.198 | -0.0222 | No | ||
| 120 | TNNI3 | 89 | 16231 | -0.246 | -0.0287 | No | ||
| 121 | HRAS | 4868 | 16569 | -0.329 | -0.0384 | No | ||
| 122 | RNH1 | 17562 | 16813 | -0.402 | -0.0411 | No | ||
| 123 | GNPAT | 18420 | 17262 | -0.583 | -0.0502 | No | ||
| 124 | BAX | 17832 | 17840 | -0.932 | -0.0572 | No | ||
| 125 | TGFB2 | 5743 | 17912 | -0.982 | -0.0356 | No | ||
| 126 | BTG1 | 4458 4457 8663 | 17932 | -0.993 | -0.0108 | No | ||
| 127 | ANGPTL4 | 1504 7178 | 18408 | -1.840 | 0.0112 | No |

