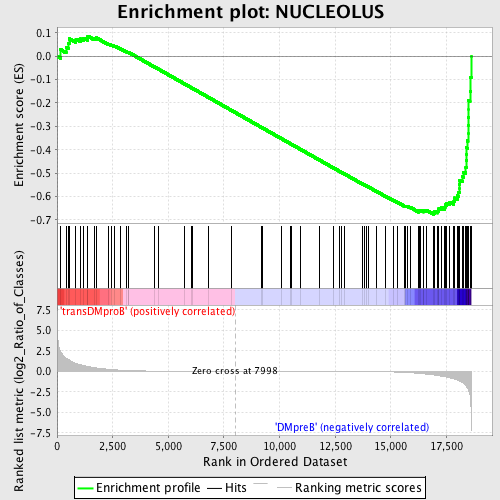
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | NUCLEOLUS |
| Enrichment Score (ES) | -0.6769384 |
| Normalized Enrichment Score (NES) | -1.5690528 |
| Nominal p-value | 0.0022421526 |
| FDR q-value | 0.113602884 |
| FWER p-Value | 0.776 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | WRN | 5882 10315 | 155 | 2.357 | 0.0281 | No | ||
| 2 | CCNT1 | 22140 11607 | 405 | 1.594 | 0.0393 | No | ||
| 3 | NOL6 | 2385 2405 15913 2466 | 508 | 1.438 | 0.0561 | No | ||
| 4 | PARP1 | 8559 14027 | 559 | 1.347 | 0.0742 | No | ||
| 5 | EXOSC10 | 2482 11943 | 848 | 0.951 | 0.0734 | No | ||
| 6 | DDX11 | 23169 | 1065 | 0.789 | 0.0739 | No | ||
| 7 | ZNF346 | 3163 6517 3193 | 1194 | 0.685 | 0.0776 | No | ||
| 8 | TP53 | 20822 | 1359 | 0.593 | 0.0779 | No | ||
| 9 | TOP1 | 5790 5789 10210 | 1379 | 0.582 | 0.0859 | No | ||
| 10 | TOP2A | 20257 | 1663 | 0.453 | 0.0777 | No | ||
| 11 | TCOF1 | 5660 | 1760 | 0.407 | 0.0788 | No | ||
| 12 | RPL11 | 12450 | 2322 | 0.239 | 0.0522 | No | ||
| 13 | DEDD2 | 7429 | 2466 | 0.207 | 0.0477 | No | ||
| 14 | MPHOSPH1 | 23878 | 2600 | 0.178 | 0.0433 | No | ||
| 15 | RPS7 | 9760 | 2839 | 0.131 | 0.0324 | No | ||
| 16 | RPL35 | 12360 | 3096 | 0.096 | 0.0201 | No | ||
| 17 | SRP68 | 20143 | 3210 | 0.084 | 0.0153 | No | ||
| 18 | ABL1 | 2693 4301 2794 | 3222 | 0.082 | 0.0160 | No | ||
| 19 | FGF18 | 4721 1351 | 4370 | 0.022 | -0.0456 | No | ||
| 20 | DDX47 | 17261 12577 1133 | 4553 | 0.018 | -0.0551 | No | ||
| 21 | UBTF | 1313 10052 | 5736 | 0.007 | -0.1188 | No | ||
| 22 | NF2 | 1222 5166 | 6031 | 0.006 | -0.1346 | No | ||
| 23 | NLRP5 | 10743 18358 | 6071 | 0.005 | -0.1366 | No | ||
| 24 | POLA1 | 24112 | 6822 | 0.003 | -0.1770 | No | ||
| 25 | APTX | 2455 7291 | 7845 | 0.000 | -0.2322 | No | ||
| 26 | FMR1 | 24321 | 9187 | -0.003 | -0.3045 | No | ||
| 27 | RBPJ | 9709 3594 5370 | 9245 | -0.003 | -0.3075 | No | ||
| 28 | DDX56 | 6984 | 10091 | -0.005 | -0.3530 | No | ||
| 29 | EXOSC9 | 15610 | 10479 | -0.006 | -0.3738 | No | ||
| 30 | MPHOSPH10 | 17805 | 10556 | -0.007 | -0.3778 | No | ||
| 31 | NPM1 | 1196 | 10938 | -0.008 | -0.3983 | No | ||
| 32 | DNAJB9 | 6565 | 11795 | -0.011 | -0.4443 | No | ||
| 33 | NEK11 | 3099 19016 | 12410 | -0.016 | -0.4772 | No | ||
| 34 | MDM2 | 19620 3327 | 12679 | -0.018 | -0.4914 | No | ||
| 35 | TOP2B | 5791 | 12798 | -0.019 | -0.4974 | No | ||
| 36 | IFI16 | 13745 | 12910 | -0.020 | -0.5031 | No | ||
| 37 | NOL4 | 2028 11432 | 13736 | -0.032 | -0.5472 | No | ||
| 38 | XRN2 | 2936 6302 2905 | 13828 | -0.033 | -0.5515 | No | ||
| 39 | PML | 3015 3074 3020 5270 | 13898 | -0.034 | -0.5547 | No | ||
| 40 | NUFIP1 | 21955 | 13995 | -0.036 | -0.5594 | No | ||
| 41 | NOL3 | 18496 915 921 | 14350 | -0.045 | -0.5778 | No | ||
| 42 | PTBP1 | 5303 | 14763 | -0.062 | -0.5990 | No | ||
| 43 | DEDD | 14056 | 15134 | -0.085 | -0.6177 | No | ||
| 44 | NCL | 5153 13899 | 15284 | -0.096 | -0.6242 | No | ||
| 45 | RPP40 | 21491 | 15629 | -0.137 | -0.6407 | No | ||
| 46 | SMUG1 | 22109 | 15671 | -0.143 | -0.6407 | No | ||
| 47 | POLR2A | 5394 | 15732 | -0.152 | -0.6416 | No | ||
| 48 | RPL3 | 11330 | 15875 | -0.177 | -0.6465 | No | ||
| 49 | SLC29A2 | 23775 | 16250 | -0.250 | -0.6628 | No | ||
| 50 | EMG1 | 1016 17005 | 16268 | -0.254 | -0.6598 | No | ||
| 51 | MORF4L2 | 12118 | 16315 | -0.265 | -0.6582 | No | ||
| 52 | DDX54 | 7778 | 16467 | -0.301 | -0.6617 | No | ||
| 53 | SURF6 | 9945 | 16490 | -0.307 | -0.6581 | No | ||
| 54 | ILF2 | 12582 | 16582 | -0.333 | -0.6579 | No | ||
| 55 | RPL36 | 12024 | 16936 | -0.449 | -0.6700 | Yes | ||
| 56 | TAF5 | 23833 5934 3759 | 16975 | -0.465 | -0.6648 | Yes | ||
| 57 | DDX24 | 2084 20995 | 17083 | -0.503 | -0.6628 | Yes | ||
| 58 | COIL | 20708 | 17131 | -0.523 | -0.6573 | Yes | ||
| 59 | KRR1 | 6989 3313 | 17161 | -0.534 | -0.6506 | Yes | ||
| 60 | ELP3 | 21782 | 17276 | -0.589 | -0.6476 | Yes | ||
| 61 | RPS19 | 5398 | 17418 | -0.653 | -0.6451 | Yes | ||
| 62 | POP7 | 16329 | 17439 | -0.665 | -0.6359 | Yes | ||
| 63 | KLHL7 | 16907 | 17507 | -0.701 | -0.6287 | Yes | ||
| 64 | RPP30 | 23877 | 17650 | -0.803 | -0.6239 | Yes | ||
| 65 | RPP38 | 14698 | 17833 | -0.930 | -0.6194 | Yes | ||
| 66 | ASNA1 | 18810 | 17860 | -0.941 | -0.6062 | Yes | ||
| 67 | NSUN2 | 21610 | 17998 | -1.071 | -0.5970 | Yes | ||
| 68 | NOLC1 | 7704 | 18044 | -1.129 | -0.5820 | Yes | ||
| 69 | NOL5A | 12474 | 18075 | -1.164 | -0.5656 | Yes | ||
| 70 | NOLA3 | 14914 | 18080 | -1.172 | -0.5477 | Yes | ||
| 71 | EXOSC1 | 12379 | 18097 | -1.193 | -0.5301 | Yes | ||
| 72 | DDX21 | 19751 | 18231 | -1.388 | -0.5158 | Yes | ||
| 73 | SDAD1 | 16479 | 18273 | -1.454 | -0.4955 | Yes | ||
| 74 | LYAR | 16856 | 18373 | -1.729 | -0.4741 | Yes | ||
| 75 | MKI67IP | 7509 | 18389 | -1.781 | -0.4474 | Yes | ||
| 76 | CIRH1A | 18478 | 18401 | -1.818 | -0.4199 | Yes | ||
| 77 | IKBKAP | 2540 6045 | 18409 | -1.841 | -0.3918 | Yes | ||
| 78 | MYBBP1A | 5217 1232 5216 | 18447 | -2.056 | -0.3620 | Yes | ||
| 79 | RRP9 | 19328 | 18469 | -2.146 | -0.3299 | Yes | ||
| 80 | POLR1E | 7226 | 18474 | -2.171 | -0.2965 | Yes | ||
| 81 | GNL3 | 21894 3356 | 18482 | -2.230 | -0.2624 | Yes | ||
| 82 | POP1 | 2214 7482 | 18497 | -2.340 | -0.2270 | Yes | ||
| 83 | NOLA2 | 11972 | 18500 | -2.357 | -0.1906 | Yes | ||
| 84 | NOL1 | 8472 17280 | 18560 | -2.904 | -0.1489 | Yes | ||
| 85 | GEMIN4 | 6592 6591 | 18592 | -3.878 | -0.0906 | Yes | ||
| 86 | CDKN2A | 2491 15841 | 18612 | -5.935 | 0.0002 | Yes |

