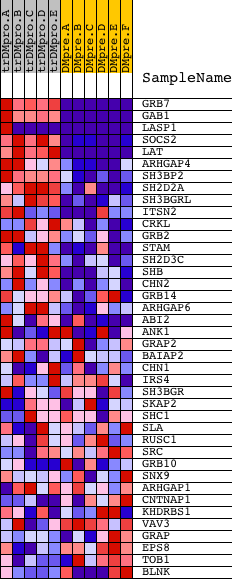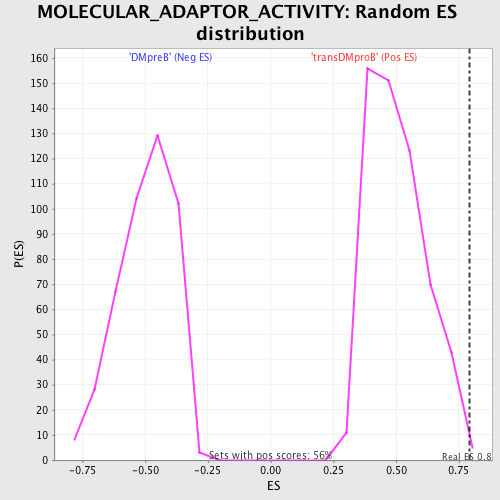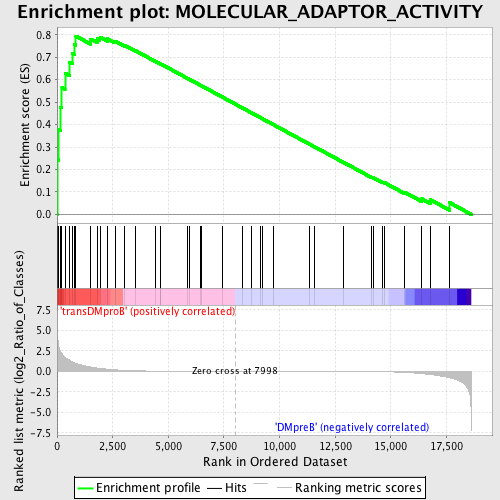
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | MOLECULAR_ADAPTOR_ACTIVITY |
| Enrichment Score (ES) | 0.79403585 |
| Normalized Enrichment Score (NES) | 1.5775135 |
| Nominal p-value | 0.0053667263 |
| FDR q-value | 0.22094846 |
| FWER p-Value | 0.66 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 5 | 5.748 | 0.2451 | Yes | ||
| 2 | GAB1 | 18828 | 64 | 3.185 | 0.3779 | Yes | ||
| 3 | LASP1 | 4986 4985 1248 | 140 | 2.442 | 0.4781 | Yes | ||
| 4 | SOCS2 | 5694 | 217 | 2.127 | 0.5648 | Yes | ||
| 5 | LAT | 17643 | 357 | 1.694 | 0.6297 | Yes | ||
| 6 | ARHGAP4 | 9343 | 548 | 1.364 | 0.6777 | Yes | ||
| 7 | SH3BP2 | 16874 | 692 | 1.119 | 0.7178 | Yes | ||
| 8 | SH2D2A | 6569 | 771 | 1.025 | 0.7573 | Yes | ||
| 9 | SH3BGRL | 24267 | 846 | 0.954 | 0.7940 | Yes | ||
| 10 | ITSN2 | 21327 | 1508 | 0.523 | 0.7808 | No | ||
| 11 | CRKL | 4560 | 1800 | 0.391 | 0.7818 | No | ||
| 12 | GRB2 | 20149 | 1927 | 0.347 | 0.7899 | No | ||
| 13 | STAM | 2912 15117 | 2270 | 0.249 | 0.7821 | No | ||
| 14 | SH2D3C | 2864 | 2605 | 0.176 | 0.7716 | No | ||
| 15 | SHB | 10493 | 3022 | 0.103 | 0.7536 | No | ||
| 16 | CHN2 | 7652 | 3536 | 0.057 | 0.7284 | No | ||
| 17 | GRB14 | 14574 2719 | 4432 | 0.021 | 0.6811 | No | ||
| 18 | ARHGAP6 | 24207 | 4639 | 0.016 | 0.6707 | No | ||
| 19 | ABI2 | 4037 6830 | 4664 | 0.016 | 0.6701 | No | ||
| 20 | ANK1 | 22501 18650 4385 8590 18649 | 5870 | 0.006 | 0.6055 | No | ||
| 21 | GRAP2 | 5113 9398 | 5964 | 0.006 | 0.6008 | No | ||
| 22 | BAIAP2 | 449 4236 | 6433 | 0.004 | 0.5757 | No | ||
| 23 | CHN1 | 4246 | 6482 | 0.004 | 0.5733 | No | ||
| 24 | IRS4 | 9183 4926 | 7412 | 0.001 | 0.5234 | No | ||
| 25 | SH3BGR | 11937 | 8311 | -0.001 | 0.4751 | No | ||
| 26 | SKAP2 | 17153 | 8741 | -0.002 | 0.4520 | No | ||
| 27 | SHC1 | 9813 9812 5430 | 9137 | -0.003 | 0.4309 | No | ||
| 28 | SLA | 5449 | 9234 | -0.003 | 0.4259 | No | ||
| 29 | RUSC1 | 1853 15283 | 9737 | -0.004 | 0.3990 | No | ||
| 30 | SRC | 5507 | 11341 | -0.009 | 0.3131 | No | ||
| 31 | GRB10 | 4799 | 11584 | -0.010 | 0.3006 | No | ||
| 32 | SNX9 | 23132 | 12859 | -0.019 | 0.2328 | No | ||
| 33 | ARHGAP1 | 6001 10448 | 14130 | -0.040 | 0.1661 | No | ||
| 34 | CNTNAP1 | 11987 | 14234 | -0.042 | 0.1624 | No | ||
| 35 | KHDRBS1 | 5405 9778 | 14646 | -0.057 | 0.1427 | No | ||
| 36 | VAV3 | 1774 1848 15443 | 14719 | -0.060 | 0.1414 | No | ||
| 37 | GRAP | 20851 | 15632 | -0.137 | 0.0982 | No | ||
| 38 | EPS8 | 16948 | 16398 | -0.284 | 0.0691 | No | ||
| 39 | TOB1 | 20703 | 16778 | -0.392 | 0.0654 | No | ||
| 40 | BLNK | 23681 3691 | 17637 | -0.784 | 0.0527 | No |