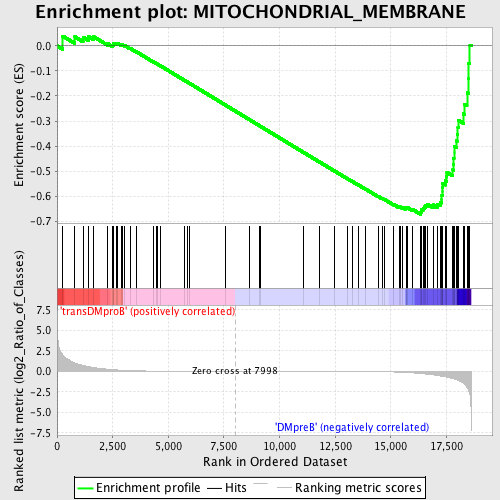
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | MITOCHONDRIAL_MEMBRANE |
| Enrichment Score (ES) | -0.6729678 |
| Normalized Enrichment Score (NES) | -1.5374253 |
| Nominal p-value | 0.004651163 |
| FDR q-value | 0.1566386 |
| FWER p-Value | 0.937 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RAB11FIP5 | 17089 | 256 | 1.998 | 0.0390 | No | ||
| 2 | CASP7 | 23826 | 800 | 0.999 | 0.0361 | No | ||
| 3 | MCL1 | 15502 | 1166 | 0.703 | 0.0349 | No | ||
| 4 | SLC25A11 | 20366 1368 | 1400 | 0.573 | 0.0375 | No | ||
| 5 | SLC25A1 | 22645 | 1650 | 0.458 | 0.0362 | No | ||
| 6 | TIMM17B | 24390 | 2283 | 0.247 | 0.0086 | No | ||
| 7 | OGDH | 9502 1412 | 2481 | 0.204 | 0.0033 | No | ||
| 8 | NDUFA13 | 7400 | 2525 | 0.195 | 0.0062 | No | ||
| 9 | UQCRB | 12545 | 2529 | 0.195 | 0.0112 | No | ||
| 10 | ALAS2 | 24229 | 2650 | 0.166 | 0.0091 | No | ||
| 11 | UQCRH | 12378 | 2701 | 0.155 | 0.0105 | No | ||
| 12 | SLC25A22 | 12671 | 2891 | 0.120 | 0.0034 | No | ||
| 13 | NDUFA6 | 12473 | 2933 | 0.114 | 0.0042 | No | ||
| 14 | RAF1 | 17035 | 3038 | 0.101 | 0.0013 | No | ||
| 15 | NDUFS4 | 9452 5157 3173 | 3285 | 0.077 | -0.0099 | No | ||
| 16 | OPA1 | 13169 22800 7894 | 3578 | 0.054 | -0.0243 | No | ||
| 17 | HSD3B2 | 4873 4874 15229 4875 9126 | 4343 | 0.023 | -0.0649 | No | ||
| 18 | HCCS | 9079 4842 | 4459 | 0.020 | -0.0706 | No | ||
| 19 | AIFM2 | 3320 3407 20006 3317 | 4505 | 0.019 | -0.0725 | No | ||
| 20 | VDAC1 | 10282 | 4635 | 0.016 | -0.0790 | No | ||
| 21 | SDHA | 12436 | 5746 | 0.007 | -0.1387 | No | ||
| 22 | MTX2 | 14980 | 5881 | 0.006 | -0.1458 | No | ||
| 23 | COX6B2 | 17983 4110 | 5951 | 0.006 | -0.1493 | No | ||
| 24 | PHB | 9558 | 7582 | 0.001 | -0.2372 | No | ||
| 25 | NDUFS1 | 13940 | 8627 | -0.001 | -0.2935 | No | ||
| 26 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.003 | -0.3197 | No | ||
| 27 | MFN2 | 15678 2417 | 9149 | -0.003 | -0.3214 | No | ||
| 28 | BNIP3 | 17581 | 11061 | -0.008 | -0.4243 | No | ||
| 29 | IMMT | 367 8029 | 11808 | -0.012 | -0.4642 | No | ||
| 30 | UCP3 | 3972 18175 | 12471 | -0.016 | -0.4995 | No | ||
| 31 | MRPL32 | 7961 | 13045 | -0.021 | -0.5298 | No | ||
| 32 | PPOX | 13761 4104 | 13267 | -0.024 | -0.5411 | No | ||
| 33 | TIMM23 | 7004 5768 | 13528 | -0.028 | -0.5544 | No | ||
| 34 | NDUFA1 | 24170 | 13863 | -0.034 | -0.5715 | No | ||
| 35 | FIS1 | 16669 3579 | 14426 | -0.048 | -0.6006 | No | ||
| 36 | HSD3B1 | 15227 | 14622 | -0.055 | -0.6096 | No | ||
| 37 | ATP5B | 19846 | 14718 | -0.060 | -0.6132 | No | ||
| 38 | NNT | 3273 5181 9471 3238 | 15128 | -0.084 | -0.6330 | No | ||
| 39 | SLC25A3 | 19644 | 15387 | -0.109 | -0.6440 | No | ||
| 40 | RHOT1 | 12227 | 15412 | -0.111 | -0.6424 | No | ||
| 41 | NDUFS8 | 23950 | 15526 | -0.123 | -0.6452 | No | ||
| 42 | ATP5C1 | 8635 | 15682 | -0.144 | -0.6498 | No | ||
| 43 | NDUFA2 | 9450 | 15685 | -0.144 | -0.6461 | No | ||
| 44 | TIMM9 | 19686 | 15736 | -0.152 | -0.6448 | No | ||
| 45 | UQCRC1 | 10259 | 15952 | -0.190 | -0.6513 | No | ||
| 46 | ATP5D | 19949 | 16354 | -0.272 | -0.6658 | Yes | ||
| 47 | TOMM34 | 14359 | 16370 | -0.277 | -0.6593 | Yes | ||
| 48 | ATP5G2 | 12610 | 16388 | -0.282 | -0.6527 | Yes | ||
| 49 | NDUFAB1 | 7667 | 16449 | -0.296 | -0.6482 | Yes | ||
| 50 | ATP5G3 | 14558 | 16502 | -0.310 | -0.6428 | Yes | ||
| 51 | SURF1 | 14645 | 16563 | -0.327 | -0.6374 | Yes | ||
| 52 | ABCB7 | 24085 | 16659 | -0.359 | -0.6330 | Yes | ||
| 53 | ATP5O | 22539 | 16907 | -0.437 | -0.6348 | Yes | ||
| 54 | NDUFS7 | 19946 | 17093 | -0.507 | -0.6314 | Yes | ||
| 55 | NDUFS2 | 4089 13760 | 17217 | -0.557 | -0.6233 | Yes | ||
| 56 | NDUFS3 | 7553 12683 | 17292 | -0.597 | -0.6115 | Yes | ||
| 57 | COX15 | 5931 | 17295 | -0.598 | -0.5958 | Yes | ||
| 58 | TOMM22 | 5863 | 17320 | -0.607 | -0.5811 | Yes | ||
| 59 | VDAC3 | 10284 | 17322 | -0.607 | -0.5651 | Yes | ||
| 60 | ATP5E | 14321 | 17331 | -0.611 | -0.5494 | Yes | ||
| 61 | ATP5A1 | 23505 | 17444 | -0.667 | -0.5378 | Yes | ||
| 62 | NDUFA9 | 12288 | 17506 | -0.699 | -0.5227 | Yes | ||
| 63 | MPV17 | 9407 3505 3543 | 17521 | -0.710 | -0.5047 | Yes | ||
| 64 | SDHD | 19125 | 17790 | -0.909 | -0.4951 | Yes | ||
| 65 | NDUFV1 | 23953 | 17810 | -0.917 | -0.4719 | Yes | ||
| 66 | BCS1L | 14223 | 17817 | -0.921 | -0.4479 | Yes | ||
| 67 | PMPCA | 12421 7350 | 17841 | -0.933 | -0.4245 | Yes | ||
| 68 | TIMM8B | 11391 | 17842 | -0.933 | -0.3999 | Yes | ||
| 69 | TIMM50 | 17911 | 17958 | -1.026 | -0.3790 | Yes | ||
| 70 | TIMM17A | 13822 | 17980 | -1.048 | -0.3524 | Yes | ||
| 71 | ATP5G1 | 8636 | 18017 | -1.094 | -0.3255 | Yes | ||
| 72 | VDAC2 | 22081 | 18042 | -1.127 | -0.2970 | Yes | ||
| 73 | ATP5J | 880 22555 | 18255 | -1.424 | -0.2708 | Yes | ||
| 74 | PHB2 | 17286 | 18315 | -1.555 | -0.2329 | Yes | ||
| 75 | RHOT2 | 1533 10073 | 18425 | -1.956 | -0.1872 | Yes | ||
| 76 | SLC25A15 | 356 3819 | 18493 | -2.279 | -0.1306 | Yes | ||
| 77 | TIMM13 | 14974 | 18499 | -2.353 | -0.0687 | Yes | ||
| 78 | TIMM10 | 11390 2158 | 18555 | -2.838 | 0.0033 | Yes |

