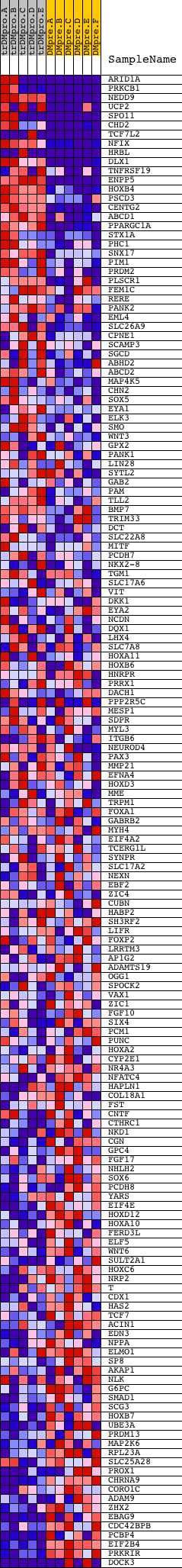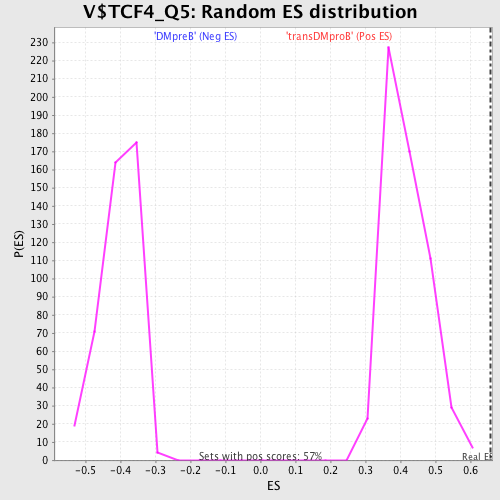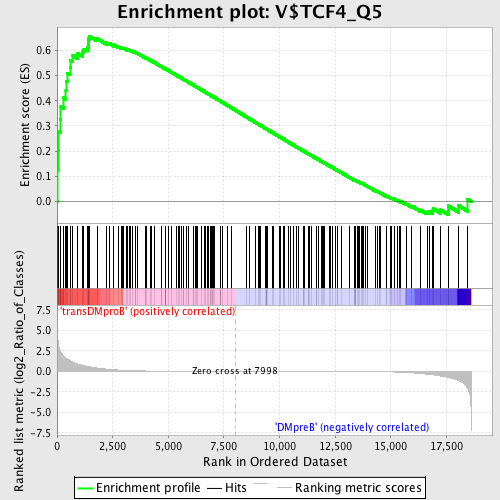
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$TCF4_Q5 |
| Enrichment Score (ES) | 0.6568953 |
| Normalized Enrichment Score (NES) | 1.5795698 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.048960175 |
| FWER p-Value | 0.35 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 6 | 5.440 | 0.1231 | Yes | ||
| 2 | PRKCB1 | 1693 9574 | 42 | 3.635 | 0.2037 | Yes | ||
| 3 | NEDD9 | 3206 21483 | 61 | 3.267 | 0.2768 | Yes | ||
| 4 | UCP2 | 5829 3059 | 152 | 2.369 | 0.3257 | Yes | ||
| 5 | SPO11 | 2666 14717 | 171 | 2.289 | 0.3767 | Yes | ||
| 6 | CHD2 | 10847 | 300 | 1.835 | 0.4114 | Yes | ||
| 7 | TCF7L2 | 10048 5646 | 397 | 1.602 | 0.4425 | Yes | ||
| 8 | NFIX | 9458 | 409 | 1.590 | 0.4780 | Yes | ||
| 9 | HRBL | 3452 16326 3614 | 453 | 1.525 | 0.5103 | Yes | ||
| 10 | DLX1 | 14988 | 581 | 1.306 | 0.5330 | Yes | ||
| 11 | TNFRSF19 | 21799 3033 | 608 | 1.257 | 0.5601 | Yes | ||
| 12 | ENPP5 | 23230 | 709 | 1.100 | 0.5797 | Yes | ||
| 13 | HOXB4 | 20686 | 912 | 0.898 | 0.5891 | Yes | ||
| 14 | PSCD3 | 16633 | 1125 | 0.736 | 0.5943 | Yes | ||
| 15 | CENTG2 | 6874 | 1204 | 0.678 | 0.6055 | Yes | ||
| 16 | ABCD1 | 24305 | 1348 | 0.599 | 0.6113 | Yes | ||
| 17 | PPARGC1A | 16533 | 1402 | 0.572 | 0.6214 | Yes | ||
| 18 | STX1A | 16679 | 1410 | 0.567 | 0.6339 | Yes | ||
| 19 | PHC1 | 17013 | 1422 | 0.560 | 0.6460 | Yes | ||
| 20 | SNX17 | 3495 16885 | 1452 | 0.549 | 0.6569 | Yes | ||
| 21 | PIM1 | 23308 | 1801 | 0.391 | 0.6469 | No | ||
| 22 | PRDM2 | 2522 4289 | 2227 | 0.260 | 0.6298 | No | ||
| 23 | PLSCR1 | 10218 | 2346 | 0.234 | 0.6287 | No | ||
| 24 | FEM1C | 23441 10771 23442 | 2556 | 0.189 | 0.6217 | No | ||
| 25 | RERE | 12722 | 2769 | 0.142 | 0.6134 | No | ||
| 26 | PANK2 | 14838 | 2896 | 0.119 | 0.6093 | No | ||
| 27 | EML4 | 8122 | 2937 | 0.113 | 0.6097 | No | ||
| 28 | SLC26A9 | 11560 980 4067 | 2986 | 0.107 | 0.6095 | No | ||
| 29 | CPNE1 | 11186 | 3138 | 0.091 | 0.6034 | No | ||
| 30 | SCAMP3 | 15543 | 3150 | 0.090 | 0.6049 | No | ||
| 31 | SGCD | 1425 10779 | 3260 | 0.079 | 0.6008 | No | ||
| 32 | ABHD2 | 18208 12043 | 3316 | 0.074 | 0.5995 | No | ||
| 33 | ABCD2 | 11204 6494 | 3397 | 0.067 | 0.5966 | No | ||
| 34 | MAP4K5 | 11853 21048 2101 2142 | 3505 | 0.059 | 0.5922 | No | ||
| 35 | CHN2 | 7652 | 3536 | 0.057 | 0.5918 | No | ||
| 36 | SOX5 | 9850 1044 5480 16937 | 3619 | 0.051 | 0.5885 | No | ||
| 37 | EYA1 | 4695 4061 | 3951 | 0.035 | 0.5714 | No | ||
| 38 | ELK3 | 4663 94 3388 | 3985 | 0.033 | 0.5704 | No | ||
| 39 | SMO | 11474 6702 | 4015 | 0.032 | 0.5695 | No | ||
| 40 | WNT3 | 20635 | 4195 | 0.026 | 0.5604 | No | ||
| 41 | GPX2 | 9037 | 4238 | 0.025 | 0.5587 | No | ||
| 42 | PANK1 | 1687 23692 | 4358 | 0.023 | 0.5528 | No | ||
| 43 | LIN28 | 15723 | 4680 | 0.016 | 0.5358 | No | ||
| 44 | SYTL2 | 18190 1539 3730 | 4694 | 0.015 | 0.5354 | No | ||
| 45 | GAB2 | 1821 18184 2025 | 4849 | 0.013 | 0.5274 | No | ||
| 46 | PAM | 5222 | 4858 | 0.013 | 0.5272 | No | ||
| 47 | TLL2 | 23680 | 4873 | 0.013 | 0.5268 | No | ||
| 48 | BMP7 | 14324 | 4885 | 0.013 | 0.5265 | No | ||
| 49 | TRIM33 | 8236 13579 15472 8237 | 5009 | 0.012 | 0.5201 | No | ||
| 50 | DCT | 4603 21723 | 5145 | 0.011 | 0.5130 | No | ||
| 51 | SLC22A8 | 23944 | 5349 | 0.009 | 0.5022 | No | ||
| 52 | MITF | 17349 | 5449 | 0.008 | 0.4970 | No | ||
| 53 | PCDH7 | 7029 3460 | 5496 | 0.008 | 0.4947 | No | ||
| 54 | NKX2-8 | 9465 | 5588 | 0.008 | 0.4900 | No | ||
| 55 | TGM1 | 21821 | 5688 | 0.007 | 0.4848 | No | ||
| 56 | SLC17A6 | 18226 | 5690 | 0.007 | 0.4849 | No | ||
| 57 | VIT | 23155 | 5819 | 0.007 | 0.4781 | No | ||
| 58 | DKK1 | 23700 | 5904 | 0.006 | 0.4737 | No | ||
| 59 | EYA2 | 2734 14729 | 5909 | 0.006 | 0.4736 | No | ||
| 60 | NCDN | 15756 2487 6471 | 6108 | 0.005 | 0.4630 | No | ||
| 61 | DQX1 | 17399 | 6114 | 0.005 | 0.4628 | No | ||
| 62 | LHX4 | 13800 | 6220 | 0.005 | 0.4573 | No | ||
| 63 | SLC7A8 | 3611 21833 | 6260 | 0.005 | 0.4553 | No | ||
| 64 | HOXA11 | 17146 | 6304 | 0.005 | 0.4530 | No | ||
| 65 | HOXB6 | 20688 | 6477 | 0.004 | 0.4438 | No | ||
| 66 | HNRPR | 13196 7909 2500 | 6621 | 0.003 | 0.4361 | No | ||
| 67 | PRRX1 | 9595 5271 | 6686 | 0.003 | 0.4327 | No | ||
| 68 | DACH1 | 4595 3371 | 6775 | 0.003 | 0.4280 | No | ||
| 69 | PPP2R5C | 6522 2147 | 6820 | 0.003 | 0.4257 | No | ||
| 70 | MESP1 | 17786 | 6903 | 0.003 | 0.4213 | No | ||
| 71 | SDPR | 14252 | 6914 | 0.003 | 0.4209 | No | ||
| 72 | MYL3 | 19291 | 6943 | 0.003 | 0.4194 | No | ||
| 73 | ITGB6 | 14581 2809 | 6977 | 0.002 | 0.4177 | No | ||
| 74 | NEUROD4 | 19582 | 7006 | 0.002 | 0.4162 | No | ||
| 75 | PAX3 | 13910 | 7016 | 0.002 | 0.4158 | No | ||
| 76 | MMP21 | 17589 | 7079 | 0.002 | 0.4125 | No | ||
| 77 | EFNA4 | 1866 15277 | 7325 | 0.002 | 0.3992 | No | ||
| 78 | HOXD3 | 9115 | 7336 | 0.002 | 0.3987 | No | ||
| 79 | MME | 15579 | 7345 | 0.002 | 0.3983 | No | ||
| 80 | TRPM1 | 10846 474 9395 494 | 7431 | 0.001 | 0.3937 | No | ||
| 81 | FOXA1 | 21060 | 7666 | 0.001 | 0.3811 | No | ||
| 82 | GABRB2 | 20922 | 7851 | 0.000 | 0.3711 | No | ||
| 83 | MYH4 | 20837 | 8516 | -0.001 | 0.3352 | No | ||
| 84 | EIF4A2 | 4660 1679 1645 | 8669 | -0.002 | 0.3270 | No | ||
| 85 | TCERG1L | 7690 | 8896 | -0.002 | 0.3148 | No | ||
| 86 | SYNPR | 22094 | 9048 | -0.003 | 0.3066 | No | ||
| 87 | SLC17A2 | 10163 5744 | 9109 | -0.003 | 0.3034 | No | ||
| 88 | NEXN | 12737 1903 | 9131 | -0.003 | 0.3024 | No | ||
| 89 | EBF2 | 21975 | 9362 | -0.003 | 0.2900 | No | ||
| 90 | ZIC4 | 5992 | 9401 | -0.003 | 0.2880 | No | ||
| 91 | CUBN | 14682 | 9424 | -0.003 | 0.2869 | No | ||
| 92 | HABP2 | 10364 23825 | 9428 | -0.003 | 0.2868 | No | ||
| 93 | SH3RF2 | 23571 | 9478 | -0.004 | 0.2842 | No | ||
| 94 | LIFR | 22515 9277 | 9698 | -0.004 | 0.2724 | No | ||
| 95 | FOXP2 | 4312 17523 8523 | 9740 | -0.004 | 0.2703 | No | ||
| 96 | LRRTM3 | 19744 | 10004 | -0.005 | 0.2562 | No | ||
| 97 | AP1G2 | 2677 21827 | 10040 | -0.005 | 0.2544 | No | ||
| 98 | ADAMTS19 | 23544 | 10190 | -0.006 | 0.2465 | No | ||
| 99 | OGG1 | 17331 | 10201 | -0.006 | 0.2460 | No | ||
| 100 | SPOCK2 | 13585 | 10420 | -0.006 | 0.2344 | No | ||
| 101 | VAX1 | 23636 | 10506 | -0.006 | 0.2299 | No | ||
| 102 | ZIC1 | 19040 | 10643 | -0.007 | 0.2227 | No | ||
| 103 | FGF10 | 4719 8962 | 10747 | -0.007 | 0.2173 | No | ||
| 104 | SIX4 | 5445 | 10827 | -0.008 | 0.2132 | No | ||
| 105 | PCM1 | 3780 5229 | 11056 | -0.008 | 0.2010 | No | ||
| 106 | PUNC | 19406 | 11095 | -0.008 | 0.1991 | No | ||
| 107 | HOXA2 | 17151 | 11113 | -0.009 | 0.1984 | No | ||
| 108 | CYP2E1 | 18025 | 11294 | -0.009 | 0.1889 | No | ||
| 109 | NR4A3 | 9473 16212 5183 | 11303 | -0.009 | 0.1886 | No | ||
| 110 | NFATC4 | 22002 | 11335 | -0.009 | 0.1872 | No | ||
| 111 | HAPLN1 | 21592 | 11434 | -0.010 | 0.1821 | No | ||
| 112 | COL18A1 | 19719 3360 | 11656 | -0.011 | 0.1703 | No | ||
| 113 | FST | 8985 | 11658 | -0.011 | 0.1705 | No | ||
| 114 | CNTF | 3741 8761 3680 3736 | 11752 | -0.011 | 0.1658 | No | ||
| 115 | CTHRC1 | 22485 | 11866 | -0.012 | 0.1599 | No | ||
| 116 | NKD1 | 8228 | 11920 | -0.012 | 0.1573 | No | ||
| 117 | CGN | 12896 12897 7701 | 11965 | -0.012 | 0.1552 | No | ||
| 118 | GPC4 | 24153 | 12024 | -0.013 | 0.1523 | No | ||
| 119 | FGF17 | 21757 | 12265 | -0.014 | 0.1397 | No | ||
| 120 | NHLH2 | 9462 | 12279 | -0.015 | 0.1393 | No | ||
| 121 | SOX6 | 3684 5481 9851 | 12375 | -0.015 | 0.1345 | No | ||
| 122 | PCDH8 | 21739 3017 | 12514 | -0.016 | 0.1274 | No | ||
| 123 | YARS | 16071 | 12601 | -0.017 | 0.1231 | No | ||
| 124 | EIF4E | 15403 1827 8890 | 12612 | -0.017 | 0.1230 | No | ||
| 125 | HOXD12 | 9113 | 12800 | -0.019 | 0.1133 | No | ||
| 126 | HOXA10 | 17145 989 | 13144 | -0.023 | 0.0952 | No | ||
| 127 | FERD3L | 21292 | 13359 | -0.025 | 0.0842 | No | ||
| 128 | ELF5 | 14936 2913 | 13369 | -0.025 | 0.0843 | No | ||
| 129 | WNT6 | 5881 | 13391 | -0.026 | 0.0837 | No | ||
| 130 | SULT2A1 | 5527 | 13396 | -0.026 | 0.0841 | No | ||
| 131 | HOXC6 | 22339 2302 | 13410 | -0.026 | 0.0839 | No | ||
| 132 | NRP2 | 5194 9486 5195 | 13510 | -0.028 | 0.0792 | No | ||
| 133 | T | 23388 | 13541 | -0.028 | 0.0782 | No | ||
| 134 | CDX1 | 23429 | 13584 | -0.029 | 0.0766 | No | ||
| 135 | HAS2 | 22293 | 13661 | -0.030 | 0.0732 | No | ||
| 136 | TCF7 | 1467 20466 | 13726 | -0.031 | 0.0704 | No | ||
| 137 | ACIN1 | 3319 2807 21832 | 13754 | -0.032 | 0.0697 | No | ||
| 138 | EDN3 | 14714 | 13867 | -0.034 | 0.0644 | No | ||
| 139 | NPPA | 298 | 13945 | -0.035 | 0.0610 | No | ||
| 140 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 14321 | -0.044 | 0.0417 | No | ||
| 141 | SP8 | 21122 | 14401 | -0.047 | 0.0385 | No | ||
| 142 | AKAP1 | 4364 1326 1286 20299 | 14487 | -0.050 | 0.0350 | No | ||
| 143 | NLK | 5179 5178 | 14530 | -0.051 | 0.0339 | No | ||
| 144 | G6PC | 20656 | 14797 | -0.064 | 0.0209 | No | ||
| 145 | SMAD1 | 18831 922 | 14826 | -0.065 | 0.0209 | No | ||
| 146 | SCG3 | 19061 | 14997 | -0.075 | 0.0134 | No | ||
| 147 | HOXB7 | 666 9110 | 15044 | -0.078 | 0.0126 | No | ||
| 148 | UBE3A | 1209 1084 1830 | 15158 | -0.086 | 0.0085 | No | ||
| 149 | PRDM13 | 15933 | 15281 | -0.096 | 0.0040 | No | ||
| 150 | MAP2K6 | 20614 1414 | 15369 | -0.106 | 0.0017 | No | ||
| 151 | RPL23A | 11193 | 15456 | -0.115 | -0.0003 | No | ||
| 152 | SLC25A28 | 23669 | 15709 | -0.149 | -0.0106 | No | ||
| 153 | PROX1 | 9623 | 15932 | -0.187 | -0.0184 | No | ||
| 154 | CHRNA9 | 16836 | 16337 | -0.269 | -0.0342 | No | ||
| 155 | CORO1C | 16425 | 16631 | -0.349 | -0.0421 | No | ||
| 156 | ADAM9 | 18910 | 16746 | -0.383 | -0.0396 | No | ||
| 157 | ZHX2 | 11842 | 16891 | -0.433 | -0.0376 | No | ||
| 158 | EBAG9 | 2237 12065 | 16897 | -0.434 | -0.0280 | No | ||
| 159 | CDC42BPB | 20982 | 17249 | -0.577 | -0.0340 | No | ||
| 160 | PCBP4 | 12235 | 17584 | -0.752 | -0.0350 | No | ||
| 161 | EIF2B4 | 16574 | 17589 | -0.755 | -0.0181 | No | ||
| 162 | PRKRIR | 7834 | 18055 | -1.148 | -0.0172 | No | ||
| 163 | DOCK3 | 9920 5537 10751 | 18459 | -2.097 | 0.0085 | No |