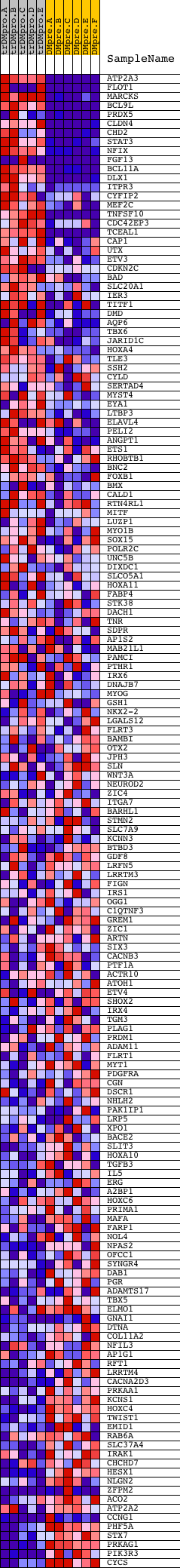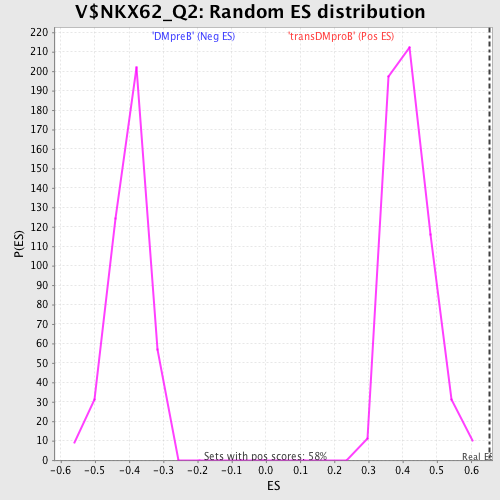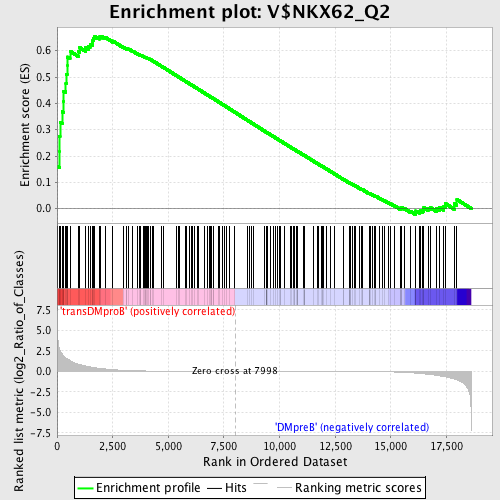
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$NKX62_Q2 |
| Enrichment Score (ES) | 0.65546584 |
| Normalized Enrichment Score (NES) | 1.5745556 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.04128288 |
| FWER p-Value | 0.379 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 20795 1314 | 0 | 7.336 | 0.1617 | Yes | ||
| 2 | FLOT1 | 23249 1613 | 105 | 2.710 | 0.2159 | Yes | ||
| 3 | MARCKS | 9331 | 110 | 2.654 | 0.2742 | Yes | ||
| 4 | BCL9L | 19475 | 131 | 2.500 | 0.3282 | Yes | ||
| 5 | PRDX5 | 7053 3732 | 230 | 2.082 | 0.3688 | Yes | ||
| 6 | CLDN4 | 8747 | 293 | 1.861 | 0.4064 | Yes | ||
| 7 | CHD2 | 10847 | 300 | 1.835 | 0.4466 | Yes | ||
| 8 | STAT3 | 5525 9906 | 396 | 1.605 | 0.4768 | Yes | ||
| 9 | NFIX | 9458 | 409 | 1.590 | 0.5112 | Yes | ||
| 10 | FGF13 | 4720 8963 2627 | 450 | 1.532 | 0.5428 | Yes | ||
| 11 | BCL11A | 4691 | 455 | 1.524 | 0.5762 | Yes | ||
| 12 | DLX1 | 14988 | 581 | 1.306 | 0.5982 | Yes | ||
| 13 | ITPR3 | 9195 | 959 | 0.861 | 0.5968 | Yes | ||
| 14 | CYFIP2 | 20485 8045 | 1018 | 0.819 | 0.6117 | Yes | ||
| 15 | MEF2C | 3204 9378 | 1283 | 0.629 | 0.6112 | Yes | ||
| 16 | TNFSF10 | 15626 | 1405 | 0.570 | 0.6172 | Yes | ||
| 17 | CDC42EP3 | 6436 | 1481 | 0.536 | 0.6250 | Yes | ||
| 18 | TCEAL1 | 24245 | 1584 | 0.487 | 0.6302 | Yes | ||
| 19 | CAP1 | 8684 | 1606 | 0.478 | 0.6396 | Yes | ||
| 20 | UTX | 10266 2574 | 1640 | 0.462 | 0.6480 | Yes | ||
| 21 | ETV3 | 11294 | 1691 | 0.439 | 0.6550 | Yes | ||
| 22 | CDKN2C | 15806 2321 | 1888 | 0.360 | 0.6523 | Yes | ||
| 23 | BAD | 24000 | 1967 | 0.335 | 0.6555 | Yes | ||
| 24 | SLC20A1 | 14855 | 2159 | 0.278 | 0.6512 | No | ||
| 25 | IER3 | 23248 | 2504 | 0.199 | 0.6370 | No | ||
| 26 | TITF1 | 10180 21062 | 2974 | 0.108 | 0.6140 | No | ||
| 27 | DMD | 24295 2647 | 3129 | 0.092 | 0.6077 | No | ||
| 28 | AQP6 | 22364 8621 | 3135 | 0.091 | 0.6094 | No | ||
| 29 | TBX6 | 1993 18075 | 3223 | 0.082 | 0.6065 | No | ||
| 30 | JARID1C | 5460 2563 | 3403 | 0.066 | 0.5982 | No | ||
| 31 | HOXA4 | 17148 | 3605 | 0.052 | 0.5885 | No | ||
| 32 | TLE3 | 5767 19416 | 3711 | 0.046 | 0.5838 | No | ||
| 33 | SSH2 | 6202 | 3751 | 0.044 | 0.5827 | No | ||
| 34 | CYLD | 18532 | 3768 | 0.043 | 0.5828 | No | ||
| 35 | SERTAD4 | 13715 | 3860 | 0.039 | 0.5787 | No | ||
| 36 | MYST4 | 7027 7026 12014 2739 | 3928 | 0.036 | 0.5759 | No | ||
| 37 | EYA1 | 4695 4061 | 3951 | 0.035 | 0.5754 | No | ||
| 38 | LTBP3 | 5008 23785 9288 | 3970 | 0.034 | 0.5752 | No | ||
| 39 | ELAVL4 | 15805 4889 9137 | 4012 | 0.032 | 0.5737 | No | ||
| 40 | PELI2 | 8217 | 4076 | 0.030 | 0.5709 | No | ||
| 41 | ANGPT1 | 8588 2260 22303 | 4099 | 0.029 | 0.5704 | No | ||
| 42 | ETS1 | 10715 6230 3135 | 4196 | 0.026 | 0.5658 | No | ||
| 43 | RHOBTB1 | 12789 | 4199 | 0.026 | 0.5662 | No | ||
| 44 | BNC2 | 15850 | 4269 | 0.025 | 0.5631 | No | ||
| 45 | FOXB1 | 12254 19069 | 4339 | 0.023 | 0.5598 | No | ||
| 46 | BMX | 2566 2585 24010 | 4683 | 0.016 | 0.5416 | No | ||
| 47 | CALD1 | 4273 8463 | 4763 | 0.015 | 0.5376 | No | ||
| 48 | RTN4RL1 | 20782 | 5375 | 0.009 | 0.5047 | No | ||
| 49 | MITF | 17349 | 5449 | 0.008 | 0.5009 | No | ||
| 50 | LUZP1 | 6533 11257 2503 | 5482 | 0.008 | 0.4994 | No | ||
| 51 | MYO1B | 13962 | 5766 | 0.007 | 0.4842 | No | ||
| 52 | SOX15 | 9848 | 5801 | 0.007 | 0.4825 | No | ||
| 53 | POLR2C | 9750 | 5939 | 0.006 | 0.4752 | No | ||
| 54 | UNC5B | 8404 | 6044 | 0.006 | 0.4697 | No | ||
| 55 | DIXDC1 | 11659 | 6065 | 0.005 | 0.4687 | No | ||
| 56 | SLCO5A1 | 6286 | 6173 | 0.005 | 0.4630 | No | ||
| 57 | HOXA11 | 17146 | 6304 | 0.005 | 0.4561 | No | ||
| 58 | FABP4 | 8601 | 6369 | 0.004 | 0.4527 | No | ||
| 59 | STK38 | 23045 | 6640 | 0.003 | 0.4382 | No | ||
| 60 | DACH1 | 4595 3371 | 6775 | 0.003 | 0.4310 | No | ||
| 61 | TNR | 5787 | 6852 | 0.003 | 0.4269 | No | ||
| 62 | SDPR | 14252 | 6914 | 0.003 | 0.4237 | No | ||
| 63 | AP1S2 | 8423 4228 | 6929 | 0.003 | 0.4230 | No | ||
| 64 | MAB21L1 | 15589 5046 | 6938 | 0.003 | 0.4226 | No | ||
| 65 | PAMCI | 19887 | 7017 | 0.002 | 0.4184 | No | ||
| 66 | PTHR1 | 5318 18985 | 7021 | 0.002 | 0.4183 | No | ||
| 67 | IRX6 | 7225 | 7236 | 0.002 | 0.4067 | No | ||
| 68 | DNAJB7 | 22196 | 7277 | 0.002 | 0.4046 | No | ||
| 69 | MYOG | 14129 | 7438 | 0.001 | 0.3960 | No | ||
| 70 | GSH1 | 16622 | 7535 | 0.001 | 0.3908 | No | ||
| 71 | NKX2-2 | 5176 | 7599 | 0.001 | 0.3874 | No | ||
| 72 | LGALS12 | 12076 | 7770 | 0.001 | 0.3782 | No | ||
| 73 | FLRT3 | 7730 12930 2673 | 7954 | 0.000 | 0.3683 | No | ||
| 74 | BAMBI | 23629 | 8540 | -0.001 | 0.3366 | No | ||
| 75 | OTX2 | 9519 | 8660 | -0.002 | 0.3302 | No | ||
| 76 | JPH3 | 18441 | 8721 | -0.002 | 0.3270 | No | ||
| 77 | SLN | 19450 | 8815 | -0.002 | 0.3220 | No | ||
| 78 | WNT3A | 20431 | 9309 | -0.003 | 0.2953 | No | ||
| 79 | NEUROD2 | 20262 | 9400 | -0.003 | 0.2905 | No | ||
| 80 | ZIC4 | 5992 | 9401 | -0.003 | 0.2906 | No | ||
| 81 | ITGA7 | 19835 | 9405 | -0.003 | 0.2905 | No | ||
| 82 | BARHL1 | 14634 | 9413 | -0.003 | 0.2902 | No | ||
| 83 | STMN2 | 15638 | 9420 | -0.003 | 0.2900 | No | ||
| 84 | SLC7A9 | 18291 1766 | 9448 | -0.003 | 0.2886 | No | ||
| 85 | KCNN3 | 15538 1913 | 9606 | -0.004 | 0.2802 | No | ||
| 86 | BTBD3 | 10457 2975 6007 6006 | 9707 | -0.004 | 0.2748 | No | ||
| 87 | GDF8 | 9411 | 9819 | -0.004 | 0.2689 | No | ||
| 88 | LRFN5 | 6213 | 9891 | -0.005 | 0.2652 | No | ||
| 89 | LRRTM3 | 19744 | 10004 | -0.005 | 0.2592 | No | ||
| 90 | FIGN | 14575 | 10020 | -0.005 | 0.2585 | No | ||
| 91 | IRS1 | 4925 | 10028 | -0.005 | 0.2582 | No | ||
| 92 | OGG1 | 17331 | 10201 | -0.006 | 0.2490 | No | ||
| 93 | C1QTNF3 | 2224 22505 | 10503 | -0.006 | 0.2329 | No | ||
| 94 | GREM1 | 14485 | 10516 | -0.007 | 0.2324 | No | ||
| 95 | ZIC1 | 19040 | 10643 | -0.007 | 0.2257 | No | ||
| 96 | ARTN | 15783 2430 2532 | 10678 | -0.007 | 0.2240 | No | ||
| 97 | SIX3 | 5444 | 10690 | -0.007 | 0.2236 | No | ||
| 98 | CACNB3 | 22373 8679 | 10777 | -0.007 | 0.2191 | No | ||
| 99 | PTF1A | 15110 | 10788 | -0.007 | 0.2187 | No | ||
| 100 | ACTR10 | 12132 2047 | 10804 | -0.007 | 0.2180 | No | ||
| 101 | ATOH1 | 4419 | 10818 | -0.007 | 0.2175 | No | ||
| 102 | ETV4 | 20212 | 11077 | -0.008 | 0.2037 | No | ||
| 103 | SHOX2 | 1802 5432 | 11110 | -0.009 | 0.2021 | No | ||
| 104 | IRX4 | 11944 | 11126 | -0.009 | 0.2015 | No | ||
| 105 | TGM3 | 10168 | 11129 | -0.009 | 0.2016 | No | ||
| 106 | PLAG1 | 7148 15949 12161 | 11523 | -0.010 | 0.1805 | No | ||
| 107 | PRDM1 | 19775 3337 | 11687 | -0.011 | 0.1719 | No | ||
| 108 | ADAM11 | 4340 1432 | 11764 | -0.011 | 0.1681 | No | ||
| 109 | FLRT1 | 11852 | 11867 | -0.012 | 0.1628 | No | ||
| 110 | MYT1 | 36 | 11911 | -0.012 | 0.1607 | No | ||
| 111 | PDGFRA | 16824 | 11924 | -0.012 | 0.1604 | No | ||
| 112 | CGN | 12896 12897 7701 | 11965 | -0.012 | 0.1585 | No | ||
| 113 | DSCR1 | 22536 1633 | 12086 | -0.013 | 0.1523 | No | ||
| 114 | NHLH2 | 9462 | 12279 | -0.015 | 0.1422 | No | ||
| 115 | PAK1IP1 | 21660 | 12468 | -0.016 | 0.1323 | No | ||
| 116 | LRP5 | 23948 9285 | 12868 | -0.019 | 0.1111 | No | ||
| 117 | XPO1 | 4172 | 12872 | -0.020 | 0.1114 | No | ||
| 118 | BACE2 | 1738 22687 1695 | 13135 | -0.022 | 0.0977 | No | ||
| 119 | SLIT3 | 20925 | 13141 | -0.022 | 0.0979 | No | ||
| 120 | HOXA10 | 17145 989 | 13144 | -0.023 | 0.0983 | No | ||
| 121 | TGFB3 | 10161 | 13172 | -0.023 | 0.0974 | No | ||
| 122 | IL5 | 20884 10220 | 13198 | -0.023 | 0.0965 | No | ||
| 123 | ERG | 1686 8915 | 13261 | -0.024 | 0.0937 | No | ||
| 124 | A2BP1 | 11212 1652 1689 | 13350 | -0.025 | 0.0895 | No | ||
| 125 | HOXC6 | 22339 2302 | 13410 | -0.026 | 0.0868 | No | ||
| 126 | PRIMA1 | 2055 20997 | 13578 | -0.029 | 0.0784 | No | ||
| 127 | MAFA | 11708 | 13684 | -0.031 | 0.0734 | No | ||
| 128 | FARP1 | 5849 10279 | 13718 | -0.031 | 0.0723 | No | ||
| 129 | NOL4 | 2028 11432 | 13736 | -0.032 | 0.0721 | No | ||
| 130 | NPAS2 | 5186 | 14021 | -0.037 | 0.0575 | No | ||
| 131 | OFCC1 | 21486 | 14046 | -0.038 | 0.0570 | No | ||
| 132 | SYNGR4 | 17825 | 14056 | -0.038 | 0.0574 | No | ||
| 133 | DAB1 | 4594 8834 | 14090 | -0.039 | 0.0564 | No | ||
| 134 | PGR | 19564 | 14193 | -0.041 | 0.0518 | No | ||
| 135 | ADAMTS17 | 598 | 14263 | -0.043 | 0.0490 | No | ||
| 136 | TBX5 | 3531 5634 | 14317 | -0.044 | 0.0471 | No | ||
| 137 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 14321 | -0.044 | 0.0479 | No | ||
| 138 | GNAI1 | 9024 | 14511 | -0.051 | 0.0388 | No | ||
| 139 | DTNA | 8870 4647 23611 1991 | 14606 | -0.055 | 0.0349 | No | ||
| 140 | COL11A2 | 23286 | 14720 | -0.060 | 0.0301 | No | ||
| 141 | NFIL3 | 21463 | 14873 | -0.068 | 0.0234 | No | ||
| 142 | AP1G1 | 4392 | 14995 | -0.074 | 0.0185 | No | ||
| 143 | RFT1 | 663 | 15163 | -0.087 | 0.0113 | No | ||
| 144 | LRRTM4 | 17405 | 15433 | -0.113 | -0.0007 | No | ||
| 145 | CACNA2D3 | 8677 | 15449 | -0.114 | 0.0010 | No | ||
| 146 | PRKAA1 | 22520 | 15480 | -0.118 | 0.0019 | No | ||
| 147 | KCNS1 | 14358 | 15483 | -0.118 | 0.0044 | No | ||
| 148 | HOXC4 | 4864 | 15620 | -0.136 | 0.0000 | No | ||
| 149 | TWIST1 | 21291 | 15865 | -0.174 | -0.0093 | No | ||
| 150 | EMID1 | 20541 1437 | 16101 | -0.217 | -0.0173 | No | ||
| 151 | RAB6A | 18173 3905 | 16123 | -0.221 | -0.0136 | No | ||
| 152 | SLC37A4 | 8994 | 16125 | -0.221 | -0.0087 | No | ||
| 153 | IRAK1 | 4916 | 16274 | -0.256 | -0.0111 | No | ||
| 154 | CHCHD7 | 16280 | 16322 | -0.266 | -0.0078 | No | ||
| 155 | HESX1 | 22070 | 16435 | -0.293 | -0.0074 | No | ||
| 156 | NLGN2 | 10117 | 16475 | -0.303 | -0.0028 | No | ||
| 157 | ZFPM2 | 22483 | 16485 | -0.305 | 0.0034 | No | ||
| 158 | ACO2 | 8527 | 16676 | -0.361 | 0.0011 | No | ||
| 159 | ATP2A2 | 4421 3481 | 16775 | -0.391 | 0.0044 | No | ||
| 160 | CCNG1 | 4492 1236 1390 | 17057 | -0.492 | -0.0000 | No | ||
| 161 | PHF5A | 22194 | 17185 | -0.544 | 0.0051 | No | ||
| 162 | STX7 | 20070 | 17382 | -0.634 | 0.0085 | No | ||
| 163 | PRKAG1 | 22135 | 17449 | -0.668 | 0.0196 | No | ||
| 164 | PIK3R3 | 5248 | 17847 | -0.935 | 0.0187 | No | ||
| 165 | CYCS | 8821 | 17971 | -1.039 | 0.0350 | No |