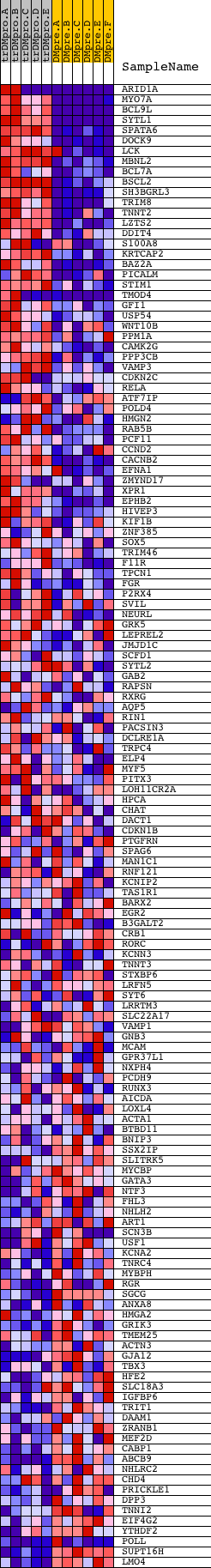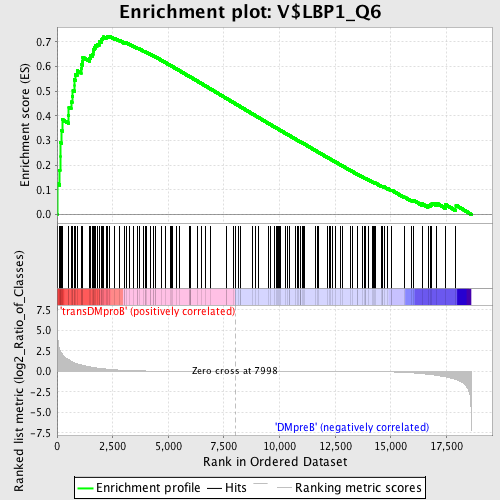
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$LBP1_Q6 |
| Enrichment Score (ES) | 0.72319394 |
| Normalized Enrichment Score (NES) | 1.7201824 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008242285 |
| FWER p-Value | 0.011 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 6 | 5.440 | 0.1244 | Yes | ||
| 2 | MYO7A | 9439 | 114 | 2.608 | 0.1784 | Yes | ||
| 3 | BCL9L | 19475 | 131 | 2.500 | 0.2349 | Yes | ||
| 4 | SYTL1 | 15728 | 134 | 2.492 | 0.2919 | Yes | ||
| 5 | SPATA6 | 16134 | 187 | 2.231 | 0.3402 | Yes | ||
| 6 | DOCK9 | 8369 | 240 | 2.049 | 0.3844 | Yes | ||
| 7 | LCK | 15746 | 521 | 1.410 | 0.4016 | Yes | ||
| 8 | MBNL2 | 21932 | 532 | 1.403 | 0.4332 | Yes | ||
| 9 | BCL7A | 8063 | 628 | 1.227 | 0.4562 | Yes | ||
| 10 | BSCL2 | 23940 | 673 | 1.151 | 0.4802 | Yes | ||
| 11 | SH3BGRL3 | 15722 | 713 | 1.093 | 0.5031 | Yes | ||
| 12 | TRIM8 | 23824 | 782 | 1.015 | 0.5227 | Yes | ||
| 13 | TNNT2 | 14113 | 784 | 1.014 | 0.5459 | Yes | ||
| 14 | LZTS2 | 10362 | 822 | 0.973 | 0.5662 | Yes | ||
| 15 | DDIT4 | 19762 | 899 | 0.908 | 0.5829 | Yes | ||
| 16 | S100A8 | 15527 | 1076 | 0.777 | 0.5912 | Yes | ||
| 17 | KRTCAP2 | 15541 | 1099 | 0.759 | 0.6074 | Yes | ||
| 18 | BAZ2A | 4371 | 1139 | 0.727 | 0.6220 | Yes | ||
| 19 | PICALM | 18191 | 1155 | 0.709 | 0.6374 | Yes | ||
| 20 | STIM1 | 18164 | 1447 | 0.550 | 0.6343 | Yes | ||
| 21 | TMOD4 | 1816 1884 15509 | 1493 | 0.530 | 0.6440 | Yes | ||
| 22 | GFI1 | 16451 | 1579 | 0.489 | 0.6507 | Yes | ||
| 23 | USP54 | 21910 | 1616 | 0.473 | 0.6595 | Yes | ||
| 24 | WNT10B | 5874 | 1629 | 0.466 | 0.6696 | Yes | ||
| 25 | PPM1A | 5279 9607 | 1677 | 0.448 | 0.6773 | Yes | ||
| 26 | CAMK2G | 21905 | 1726 | 0.420 | 0.6844 | Yes | ||
| 27 | PPP3CB | 5285 | 1794 | 0.394 | 0.6898 | Yes | ||
| 28 | VAMP3 | 15660 | 1886 | 0.361 | 0.6931 | Yes | ||
| 29 | CDKN2C | 15806 2321 | 1888 | 0.360 | 0.7013 | Yes | ||
| 30 | RELA | 23783 | 1985 | 0.329 | 0.7037 | Yes | ||
| 31 | ATF7IP | 12028 13038 | 1986 | 0.329 | 0.7112 | Yes | ||
| 32 | POLD4 | 12822 | 2043 | 0.311 | 0.7153 | Yes | ||
| 33 | HMGN2 | 9095 | 2085 | 0.298 | 0.7199 | Yes | ||
| 34 | RAB5B | 359 19591 3430 | 2204 | 0.264 | 0.7196 | Yes | ||
| 35 | PCF11 | 121 | 2247 | 0.256 | 0.7232 | Yes | ||
| 36 | CCND2 | 16987 | 2354 | 0.232 | 0.7228 | No | ||
| 37 | CACNB2 | 4466 8678 | 2590 | 0.180 | 0.7142 | No | ||
| 38 | EFNA1 | 15279 | 2793 | 0.137 | 0.7064 | No | ||
| 39 | ZMYND17 | 21909 | 3013 | 0.104 | 0.6969 | No | ||
| 40 | XPR1 | 5383 | 3023 | 0.103 | 0.6988 | No | ||
| 41 | EPHB2 | 4675 2440 8910 | 3107 | 0.095 | 0.6965 | No | ||
| 42 | HIVEP3 | 4973 | 3247 | 0.080 | 0.6908 | No | ||
| 43 | KIF1B | 15671 2476 4950 15673 | 3421 | 0.065 | 0.6829 | No | ||
| 44 | ZNF385 | 22106 2297 | 3603 | 0.052 | 0.6743 | No | ||
| 45 | SOX5 | 9850 1044 5480 16937 | 3619 | 0.051 | 0.6747 | No | ||
| 46 | TRIM46 | 1779 15281 | 3719 | 0.045 | 0.6704 | No | ||
| 47 | F11R | 9199 | 3896 | 0.038 | 0.6617 | No | ||
| 48 | TPCN1 | 16398 6383 10915 | 3962 | 0.034 | 0.6590 | No | ||
| 49 | FGR | 4723 | 3971 | 0.034 | 0.6593 | No | ||
| 50 | P2RX4 | 3520 16711 | 4008 | 0.033 | 0.6581 | No | ||
| 51 | SVIL | 1990 1942 10334 1976 23626 1946 1945 2041 | 4208 | 0.026 | 0.6479 | No | ||
| 52 | NEURL | 5164 | 4213 | 0.026 | 0.6483 | No | ||
| 53 | GRK5 | 9036 23814 | 4329 | 0.023 | 0.6426 | No | ||
| 54 | LEPREL2 | 17001 | 4352 | 0.023 | 0.6420 | No | ||
| 55 | JMJD1C | 11531 19996 | 4413 | 0.021 | 0.6392 | No | ||
| 56 | SCFD1 | 21278 2157 | 4437 | 0.020 | 0.6384 | No | ||
| 57 | SYTL2 | 18190 1539 3730 | 4694 | 0.015 | 0.6249 | No | ||
| 58 | GAB2 | 1821 18184 2025 | 4849 | 0.013 | 0.6169 | No | ||
| 59 | RAPSN | 2879 14951 | 5089 | 0.011 | 0.6042 | No | ||
| 60 | RXRG | 14063 | 5129 | 0.011 | 0.6023 | No | ||
| 61 | AQP5 | 22365 | 5168 | 0.011 | 0.6005 | No | ||
| 62 | RIN1 | 10349 | 5374 | 0.009 | 0.5896 | No | ||
| 63 | PACSIN3 | 14948 8149 2776 2665 | 5506 | 0.008 | 0.5827 | No | ||
| 64 | DCLRE1A | 23641 | 5945 | 0.006 | 0.5591 | No | ||
| 65 | TRPC4 | 15593 | 5947 | 0.006 | 0.5592 | No | ||
| 66 | ELP4 | 8094 | 5990 | 0.006 | 0.5571 | No | ||
| 67 | MYF5 | 19632 | 6004 | 0.006 | 0.5565 | No | ||
| 68 | PITX3 | 9571 | 6008 | 0.006 | 0.5565 | No | ||
| 69 | LOH11CR2A | 19498 | 6290 | 0.005 | 0.5414 | No | ||
| 70 | HPCA | 9118 | 6486 | 0.004 | 0.5309 | No | ||
| 71 | CHAT | 21882 | 6666 | 0.003 | 0.5213 | No | ||
| 72 | DACT1 | 7203 | 6904 | 0.003 | 0.5085 | No | ||
| 73 | CDKN1B | 4512 8730 | 7613 | 0.001 | 0.4702 | No | ||
| 74 | PTGFRN | 15224 | 7620 | 0.001 | 0.4699 | No | ||
| 75 | SPAG6 | 22650 | 7920 | 0.000 | 0.4537 | No | ||
| 76 | MAN1C1 | 10514 6064 15714 | 8031 | -0.000 | 0.4478 | No | ||
| 77 | RNF121 | 7952 373 | 8036 | -0.000 | 0.4476 | No | ||
| 78 | KCNIP2 | 23655 3725 | 8158 | -0.000 | 0.4410 | No | ||
| 79 | TAS1R1 | 15657 | 8221 | -0.001 | 0.4377 | No | ||
| 80 | BARX2 | 8649 | 8237 | -0.001 | 0.4369 | No | ||
| 81 | EGR2 | 8886 | 8783 | -0.002 | 0.4074 | No | ||
| 82 | B3GALT2 | 6498 | 8894 | -0.002 | 0.4015 | No | ||
| 83 | CRB1 | 5031 | 9063 | -0.003 | 0.3925 | No | ||
| 84 | RORC | 9732 | 9494 | -0.004 | 0.3693 | No | ||
| 85 | KCNN3 | 15538 1913 | 9606 | -0.004 | 0.3634 | No | ||
| 86 | TNNT3 | 18004 | 9752 | -0.004 | 0.3556 | No | ||
| 87 | STXBP6 | 21076 | 9859 | -0.005 | 0.3500 | No | ||
| 88 | LRFN5 | 6213 | 9891 | -0.005 | 0.3484 | No | ||
| 89 | SYT6 | 13437 7042 12040 | 9928 | -0.005 | 0.3466 | No | ||
| 90 | LRRTM3 | 19744 | 10004 | -0.005 | 0.3426 | No | ||
| 91 | SLC22A17 | 21830 3096 | 10052 | -0.005 | 0.3402 | No | ||
| 92 | VAMP1 | 5846 1043 | 10251 | -0.006 | 0.3296 | No | ||
| 93 | GNB3 | 9027 | 10333 | -0.006 | 0.3254 | No | ||
| 94 | MCAM | 3109 19479 | 10423 | -0.006 | 0.3207 | No | ||
| 95 | GPR37L1 | 13825 | 10459 | -0.006 | 0.3190 | No | ||
| 96 | NXPH4 | 19602 | 10696 | -0.007 | 0.3063 | No | ||
| 97 | PCDH9 | 21736 | 10825 | -0.007 | 0.2996 | No | ||
| 98 | RUNX3 | 8702 | 10854 | -0.008 | 0.2982 | No | ||
| 99 | AICDA | 17295 1087 | 10917 | -0.008 | 0.2951 | No | ||
| 100 | LOXL4 | 23672 | 10929 | -0.008 | 0.2946 | No | ||
| 101 | ACTA1 | 18715 | 10946 | -0.008 | 0.2940 | No | ||
| 102 | BTBD11 | 13147 | 11039 | -0.008 | 0.2892 | No | ||
| 103 | BNIP3 | 17581 | 11061 | -0.008 | 0.2882 | No | ||
| 104 | SSX2IP | 13617 | 11136 | -0.009 | 0.2844 | No | ||
| 105 | SLITRK5 | 21939 | 11603 | -0.011 | 0.2594 | No | ||
| 106 | MYCBP | 7093 | 11721 | -0.011 | 0.2533 | No | ||
| 107 | GATA3 | 9004 | 11750 | -0.011 | 0.2521 | No | ||
| 108 | NTF3 | 16991 | 12165 | -0.014 | 0.2300 | No | ||
| 109 | FHL3 | 16092 2457 | 12233 | -0.014 | 0.2267 | No | ||
| 110 | NHLH2 | 9462 | 12279 | -0.015 | 0.2246 | No | ||
| 111 | ART1 | 660 4414 | 12394 | -0.016 | 0.2188 | No | ||
| 112 | SCN3B | 39 3117 6163 10646 | 12525 | -0.016 | 0.2121 | No | ||
| 113 | USF1 | 5832 10260 10261 | 12726 | -0.018 | 0.2017 | No | ||
| 114 | KCNA2 | 15458 | 12811 | -0.019 | 0.1976 | No | ||
| 115 | TNRC4 | 15514 | 13167 | -0.023 | 0.1789 | No | ||
| 116 | MYBPH | 14130 | 13256 | -0.024 | 0.1747 | No | ||
| 117 | RGR | 21874 3042 | 13483 | -0.027 | 0.1631 | No | ||
| 118 | SGCG | 21798 | 13502 | -0.028 | 0.1627 | No | ||
| 119 | ANXA8 | 22046 | 13706 | -0.031 | 0.1525 | No | ||
| 120 | HMGA2 | 4857 9098 3341 3444 | 13837 | -0.033 | 0.1462 | No | ||
| 121 | GRIK3 | 16083 | 13849 | -0.034 | 0.1464 | No | ||
| 122 | TMEM25 | 2992 19141 | 13855 | -0.034 | 0.1469 | No | ||
| 123 | ACTN3 | 23967 8540 | 14013 | -0.037 | 0.1392 | No | ||
| 124 | GJA12 | 20433 1323 | 14189 | -0.041 | 0.1307 | No | ||
| 125 | TBX3 | 16723 | 14201 | -0.041 | 0.1310 | No | ||
| 126 | HFE2 | 1777 15495 | 14286 | -0.043 | 0.1275 | No | ||
| 127 | SLC18A3 | 21883 | 14303 | -0.044 | 0.1276 | No | ||
| 128 | IGFBP6 | 9161 | 14582 | -0.054 | 0.1138 | No | ||
| 129 | TRIT1 | 16098 | 14610 | -0.055 | 0.1136 | No | ||
| 130 | DAAM1 | 5536 | 14638 | -0.057 | 0.1134 | No | ||
| 131 | ZRANB1 | 6884 11702 | 14697 | -0.059 | 0.1117 | No | ||
| 132 | MEF2D | 9379 | 14867 | -0.067 | 0.1041 | No | ||
| 133 | CABP1 | 16412 | 15010 | -0.076 | 0.0981 | No | ||
| 134 | ABCB9 | 3576 3546 7096 | 15036 | -0.077 | 0.0985 | No | ||
| 135 | NHLRC2 | 23828 | 15609 | -0.134 | 0.0706 | No | ||
| 136 | CHD4 | 8418 17281 4225 | 15913 | -0.183 | 0.0584 | No | ||
| 137 | PRICKLE1 | 8379 | 16026 | -0.203 | 0.0570 | No | ||
| 138 | DPP3 | 7954 3746 | 16406 | -0.285 | 0.0430 | No | ||
| 139 | TNNI2 | 18005 | 16672 | -0.360 | 0.0370 | No | ||
| 140 | EIF4G2 | 1908 8892 | 16768 | -0.389 | 0.0407 | No | ||
| 141 | YTHDF2 | 5623 5624 | 16848 | -0.414 | 0.0460 | No | ||
| 142 | POLL | 23658 3688 | 17051 | -0.489 | 0.0463 | No | ||
| 143 | SUPT16H | 8541 | 17466 | -0.681 | 0.0395 | No | ||
| 144 | LMO4 | 15151 | 17928 | -0.992 | 0.0372 | No |