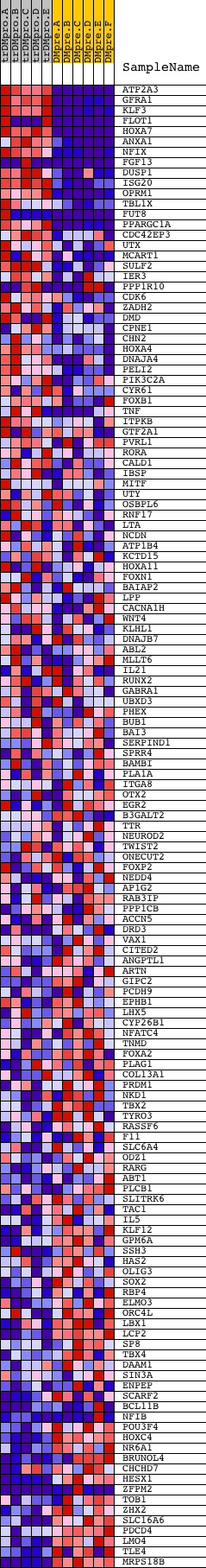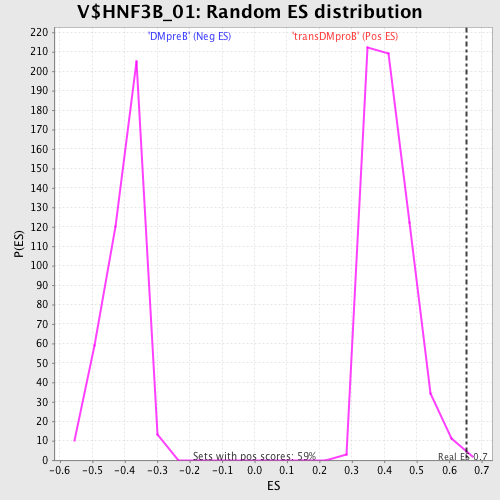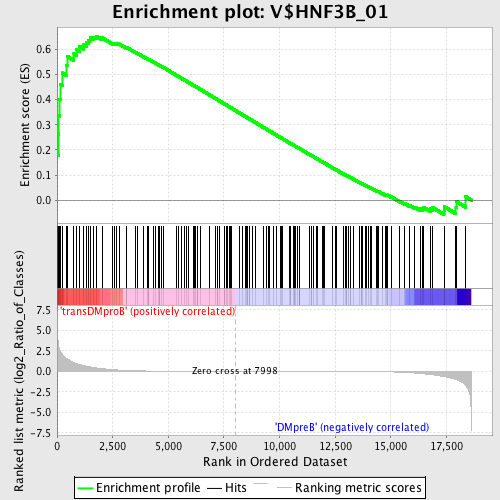
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$HNF3B_01 |
| Enrichment Score (ES) | 0.6530242 |
| Normalized Enrichment Score (NES) | 1.5732758 |
| Nominal p-value | 0.0033726813 |
| FDR q-value | 0.039816972 |
| FWER p-Value | 0.383 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 20795 1314 | 0 | 7.336 | 0.1818 | Yes | ||
| 2 | GFRA1 | 9015 | 56 | 3.423 | 0.2637 | Yes | ||
| 3 | KLF3 | 4961 | 82 | 2.952 | 0.3355 | Yes | ||
| 4 | FLOT1 | 23249 1613 | 105 | 2.710 | 0.4015 | Yes | ||
| 5 | HOXA7 | 4862 | 133 | 2.493 | 0.4618 | Yes | ||
| 6 | ANXA1 | 9282 | 243 | 2.034 | 0.5064 | Yes | ||
| 7 | NFIX | 9458 | 409 | 1.590 | 0.5368 | Yes | ||
| 8 | FGF13 | 4720 8963 2627 | 450 | 1.532 | 0.5726 | Yes | ||
| 9 | DUSP1 | 23061 | 755 | 1.046 | 0.5821 | Yes | ||
| 10 | ISG20 | 18210 | 869 | 0.933 | 0.5991 | Yes | ||
| 11 | OPRM1 | 5212 | 1014 | 0.821 | 0.6117 | Yes | ||
| 12 | TBL1X | 5628 | 1203 | 0.679 | 0.6183 | Yes | ||
| 13 | FUT8 | 2163 21233 | 1311 | 0.617 | 0.6278 | Yes | ||
| 14 | PPARGC1A | 16533 | 1402 | 0.572 | 0.6371 | Yes | ||
| 15 | CDC42EP3 | 6436 | 1481 | 0.536 | 0.6462 | Yes | ||
| 16 | UTX | 10266 2574 | 1640 | 0.462 | 0.6491 | Yes | ||
| 17 | MCART1 | 10492 | 1756 | 0.410 | 0.6530 | Yes | ||
| 18 | SULF2 | 12988 | 2034 | 0.314 | 0.6458 | No | ||
| 19 | IER3 | 23248 | 2504 | 0.199 | 0.6254 | No | ||
| 20 | PPP1R10 | 6972 11960 6971 | 2593 | 0.179 | 0.6250 | No | ||
| 21 | CDK6 | 16600 | 2683 | 0.159 | 0.6242 | No | ||
| 22 | ZADH2 | 23501 | 2791 | 0.137 | 0.6218 | No | ||
| 23 | DMD | 24295 2647 | 3129 | 0.092 | 0.6058 | No | ||
| 24 | CPNE1 | 11186 | 3138 | 0.091 | 0.6076 | No | ||
| 25 | CHN2 | 7652 | 3536 | 0.057 | 0.5876 | No | ||
| 26 | HOXA4 | 17148 | 3605 | 0.052 | 0.5852 | No | ||
| 27 | DNAJA4 | 19446 | 3898 | 0.038 | 0.5703 | No | ||
| 28 | PELI2 | 8217 | 4076 | 0.030 | 0.5615 | No | ||
| 29 | PIK3C2A | 17667 | 4095 | 0.029 | 0.5612 | No | ||
| 30 | CYR61 | 9159 15145 9311 5024 | 4127 | 0.029 | 0.5602 | No | ||
| 31 | FOXB1 | 12254 19069 | 4339 | 0.023 | 0.5494 | No | ||
| 32 | TNF | 23004 | 4411 | 0.021 | 0.5461 | No | ||
| 33 | ITPKB | 3963 14028 | 4534 | 0.018 | 0.5399 | No | ||
| 34 | GTF2A1 | 2064 8187 8188 13512 | 4590 | 0.017 | 0.5374 | No | ||
| 35 | PVRL1 | 3006 19482 12211 7192 | 4605 | 0.017 | 0.5370 | No | ||
| 36 | RORA | 3019 5388 | 4700 | 0.015 | 0.5323 | No | ||
| 37 | CALD1 | 4273 8463 | 4763 | 0.015 | 0.5293 | No | ||
| 38 | IBSP | 16775 | 5360 | 0.009 | 0.4973 | No | ||
| 39 | MITF | 17349 | 5449 | 0.008 | 0.4927 | No | ||
| 40 | UTY | 5836 | 5583 | 0.008 | 0.4857 | No | ||
| 41 | OSBPL6 | 8277 | 5738 | 0.007 | 0.4776 | No | ||
| 42 | RNF17 | 11389 | 5802 | 0.007 | 0.4743 | No | ||
| 43 | LTA | 23003 | 5892 | 0.006 | 0.4696 | No | ||
| 44 | NCDN | 15756 2487 6471 | 6108 | 0.005 | 0.4581 | No | ||
| 45 | ATP1B4 | 12586 | 6162 | 0.005 | 0.4554 | No | ||
| 46 | KCTD15 | 17864 | 6212 | 0.005 | 0.4529 | No | ||
| 47 | HOXA11 | 17146 | 6304 | 0.005 | 0.4481 | No | ||
| 48 | FOXN1 | 20337 | 6317 | 0.005 | 0.4475 | No | ||
| 49 | BAIAP2 | 449 4236 | 6433 | 0.004 | 0.4414 | No | ||
| 50 | LPP | 5570 | 6857 | 0.003 | 0.4186 | No | ||
| 51 | CACNA1H | 23077 | 7106 | 0.002 | 0.4052 | No | ||
| 52 | WNT4 | 16025 | 7201 | 0.002 | 0.4002 | No | ||
| 53 | KLHL1 | 21735 | 7211 | 0.002 | 0.3997 | No | ||
| 54 | DNAJB7 | 22196 | 7277 | 0.002 | 0.3962 | No | ||
| 55 | ABL2 | 8509 | 7508 | 0.001 | 0.3838 | No | ||
| 56 | MLLT6 | 20678 | 7530 | 0.001 | 0.3827 | No | ||
| 57 | IL21 | 12241 | 7600 | 0.001 | 0.3790 | No | ||
| 58 | RUNX2 | 4480 8700 | 7651 | 0.001 | 0.3763 | No | ||
| 59 | GABRA1 | 20492 | 7743 | 0.001 | 0.3714 | No | ||
| 60 | UBXD3 | 15701 | 7805 | 0.000 | 0.3681 | No | ||
| 61 | PHEX | 24024 | 7827 | 0.000 | 0.3670 | No | ||
| 62 | BUB1 | 8665 | 8213 | -0.001 | 0.3462 | No | ||
| 63 | BAI3 | 5580 | 8325 | -0.001 | 0.3402 | No | ||
| 64 | SERPIND1 | 22838 | 8475 | -0.001 | 0.3321 | No | ||
| 65 | SPRR4 | 15264 | 8498 | -0.001 | 0.3310 | No | ||
| 66 | BAMBI | 23629 | 8540 | -0.001 | 0.3288 | No | ||
| 67 | PLA1A | 1737 22594 | 8571 | -0.001 | 0.3272 | No | ||
| 68 | ITGA8 | 14685 | 8651 | -0.002 | 0.3229 | No | ||
| 69 | OTX2 | 9519 | 8660 | -0.002 | 0.3226 | No | ||
| 70 | EGR2 | 8886 | 8783 | -0.002 | 0.3160 | No | ||
| 71 | B3GALT2 | 6498 | 8894 | -0.002 | 0.3101 | No | ||
| 72 | TTR | 23615 | 9277 | -0.003 | 0.2895 | No | ||
| 73 | NEUROD2 | 20262 | 9400 | -0.003 | 0.2830 | No | ||
| 74 | TWIST2 | 14185 | 9516 | -0.004 | 0.2768 | No | ||
| 75 | ONECUT2 | 10346 | 9540 | -0.004 | 0.2757 | No | ||
| 76 | FOXP2 | 4312 17523 8523 | 9740 | -0.004 | 0.2650 | No | ||
| 77 | NEDD4 | 19384 3101 | 9855 | -0.005 | 0.2590 | No | ||
| 78 | AP1G2 | 2677 21827 | 10040 | -0.005 | 0.2491 | No | ||
| 79 | RAB3IP | 19623 | 10070 | -0.005 | 0.2477 | No | ||
| 80 | PPP1CB | 5282 3529 3504 | 10127 | -0.005 | 0.2448 | No | ||
| 81 | ACCN5 | 15567 | 10450 | -0.006 | 0.2275 | No | ||
| 82 | DRD3 | 22750 | 10469 | -0.006 | 0.2267 | No | ||
| 83 | VAX1 | 23636 | 10506 | -0.006 | 0.2249 | No | ||
| 84 | CITED2 | 5118 14477 | 10610 | -0.007 | 0.2195 | No | ||
| 85 | ANGPTL1 | 10195 8589 13064 | 10669 | -0.007 | 0.2165 | No | ||
| 86 | ARTN | 15783 2430 2532 | 10678 | -0.007 | 0.2163 | No | ||
| 87 | GIPC2 | 15137 | 10698 | -0.007 | 0.2154 | No | ||
| 88 | PCDH9 | 21736 | 10825 | -0.007 | 0.2088 | No | ||
| 89 | EPHB1 | 19024 | 10882 | -0.008 | 0.2059 | No | ||
| 90 | LHX5 | 9274 | 10895 | -0.008 | 0.2055 | No | ||
| 91 | CYP26B1 | 17093 | 10901 | -0.008 | 0.2054 | No | ||
| 92 | NFATC4 | 22002 | 11335 | -0.009 | 0.1822 | No | ||
| 93 | TNMD | 24260 | 11443 | -0.010 | 0.1767 | No | ||
| 94 | FOXA2 | 14405 2889 | 11453 | -0.010 | 0.1764 | No | ||
| 95 | PLAG1 | 7148 15949 12161 | 11523 | -0.010 | 0.1729 | No | ||
| 96 | COL13A1 | 8763 | 11638 | -0.011 | 0.1670 | No | ||
| 97 | PRDM1 | 19775 3337 | 11687 | -0.011 | 0.1647 | No | ||
| 98 | NKD1 | 8228 | 11920 | -0.012 | 0.1524 | No | ||
| 99 | TBX2 | 20720 | 11979 | -0.013 | 0.1496 | No | ||
| 100 | TYRO3 | 5811 | 12021 | -0.013 | 0.1477 | No | ||
| 101 | RASSF6 | 16484 | 12360 | -0.015 | 0.1298 | No | ||
| 102 | F11 | 3886 18886 | 12490 | -0.016 | 0.1232 | No | ||
| 103 | SLC6A4 | 20770 | 12564 | -0.017 | 0.1197 | No | ||
| 104 | ODZ1 | 24168 | 12857 | -0.019 | 0.1043 | No | ||
| 105 | RARG | 22113 | 12941 | -0.020 | 0.1004 | No | ||
| 106 | ABT1 | 21524 | 12997 | -0.021 | 0.0979 | No | ||
| 107 | PLCB1 | 14832 2821 | 13010 | -0.021 | 0.0978 | No | ||
| 108 | SLITRK6 | 21724 | 13094 | -0.022 | 0.0938 | No | ||
| 109 | TAC1 | 17526 19853 | 13169 | -0.023 | 0.0904 | No | ||
| 110 | IL5 | 20884 10220 | 13198 | -0.023 | 0.0894 | No | ||
| 111 | KLF12 | 4960 2637 21732 9228 | 13305 | -0.025 | 0.0843 | No | ||
| 112 | GPM6A | 3834 10618 | 13586 | -0.029 | 0.0699 | No | ||
| 113 | SSH3 | 3709 10890 | 13660 | -0.030 | 0.0667 | No | ||
| 114 | HAS2 | 22293 | 13661 | -0.030 | 0.0674 | No | ||
| 115 | OLIG3 | 20084 | 13737 | -0.032 | 0.0641 | No | ||
| 116 | SOX2 | 9849 15612 | 13875 | -0.034 | 0.0576 | No | ||
| 117 | RBP4 | 23686 | 13885 | -0.034 | 0.0579 | No | ||
| 118 | ELMO3 | 10629 18495 | 14010 | -0.037 | 0.0521 | No | ||
| 119 | ORC4L | 11172 6460 | 14092 | -0.039 | 0.0487 | No | ||
| 120 | LBX1 | 98 23659 | 14146 | -0.040 | 0.0468 | No | ||
| 121 | LCP2 | 4988 9268 | 14343 | -0.045 | 0.0373 | No | ||
| 122 | SP8 | 21122 | 14401 | -0.047 | 0.0354 | No | ||
| 123 | TBX4 | 5633 | 14442 | -0.048 | 0.0344 | No | ||
| 124 | DAAM1 | 5536 | 14638 | -0.057 | 0.0253 | No | ||
| 125 | SIN3A | 5442 | 14777 | -0.063 | 0.0194 | No | ||
| 126 | ENPEP | 4668 | 14795 | -0.064 | 0.0200 | No | ||
| 127 | SCARF2 | 22833 | 14829 | -0.065 | 0.0199 | No | ||
| 128 | BCL11B | 7190 | 14847 | -0.066 | 0.0206 | No | ||
| 129 | NFIB | 15855 | 15024 | -0.077 | 0.0129 | No | ||
| 130 | POU3F4 | 347 | 15404 | -0.110 | -0.0048 | No | ||
| 131 | HOXC4 | 4864 | 15620 | -0.136 | -0.0131 | No | ||
| 132 | NR6A1 | 14596 | 15857 | -0.173 | -0.0216 | No | ||
| 133 | BRUNOL4 | 2010 4229 | 16067 | -0.211 | -0.0277 | No | ||
| 134 | CHCHD7 | 16280 | 16322 | -0.266 | -0.0348 | No | ||
| 135 | HESX1 | 22070 | 16435 | -0.293 | -0.0336 | No | ||
| 136 | ZFPM2 | 22483 | 16485 | -0.305 | -0.0287 | No | ||
| 137 | TOB1 | 20703 | 16778 | -0.392 | -0.0348 | No | ||
| 138 | ZHX2 | 11842 | 16891 | -0.433 | -0.0301 | No | ||
| 139 | SLC16A6 | 4186 | 17391 | -0.638 | -0.0413 | No | ||
| 140 | PDCD4 | 5232 23816 | 17403 | -0.647 | -0.0259 | No | ||
| 141 | LMO4 | 15151 | 17928 | -0.992 | -0.0297 | No | ||
| 142 | TLE4 | 23720 3697 | 17948 | -1.011 | -0.0056 | No | ||
| 143 | MRPS18B | 7365 | 18359 | -1.684 | 0.0139 | No |