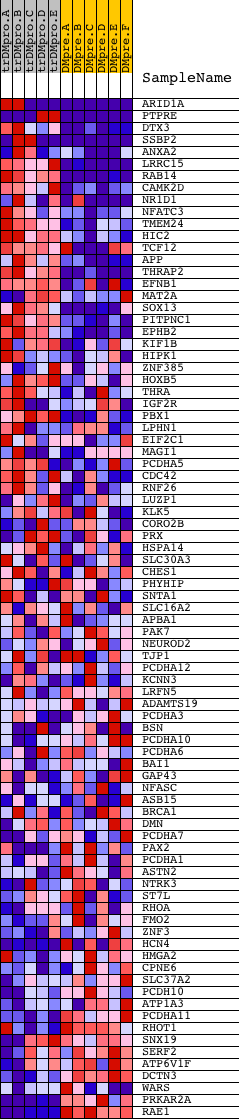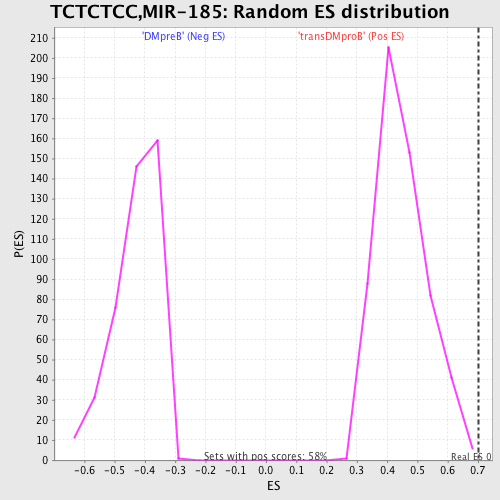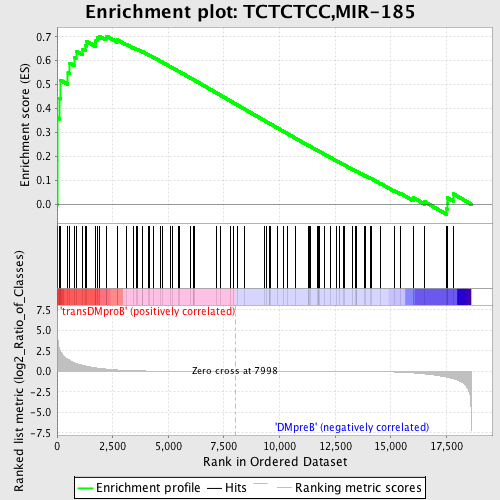
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | TCTCTCC,MIR-185 |
| Enrichment Score (ES) | 0.7027719 |
| Normalized Enrichment Score (NES) | 1.564794 |
| Nominal p-value | 0.0034722222 |
| FDR q-value | 0.042443406 |
| FWER p-Value | 0.438 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 6 | 5.440 | 0.1812 | Yes | ||
| 2 | PTPRE | 1662 18039 | 7 | 5.433 | 0.3625 | Yes | ||
| 3 | DTX3 | 19607 | 124 | 2.555 | 0.4415 | Yes | ||
| 4 | SSBP2 | 3171 7364 3252 | 153 | 2.365 | 0.5189 | Yes | ||
| 5 | ANXA2 | 19389 | 488 | 1.464 | 0.5497 | Yes | ||
| 6 | LRRC15 | 22617 | 573 | 1.327 | 0.5894 | Yes | ||
| 7 | RAB14 | 12685 | 764 | 1.035 | 0.6137 | Yes | ||
| 8 | CAMK2D | 4232 | 853 | 0.946 | 0.6405 | Yes | ||
| 9 | NR1D1 | 4650 20258 949 | 1149 | 0.715 | 0.6485 | Yes | ||
| 10 | NFATC3 | 5169 | 1280 | 0.631 | 0.6625 | Yes | ||
| 11 | TMEM24 | 19150 | 1306 | 0.619 | 0.6818 | Yes | ||
| 12 | HIC2 | 7185 | 1717 | 0.425 | 0.6739 | Yes | ||
| 13 | TCF12 | 10044 3125 | 1741 | 0.416 | 0.6865 | Yes | ||
| 14 | APP | 4402 | 1813 | 0.385 | 0.6955 | Yes | ||
| 15 | THRAP2 | 8009 | 1900 | 0.356 | 0.7028 | Yes | ||
| 16 | EFNB1 | 24285 | 2198 | 0.267 | 0.6957 | No | ||
| 17 | MAT2A | 10553 10554 6099 | 2233 | 0.259 | 0.7025 | No | ||
| 18 | SOX13 | 13835 | 2692 | 0.158 | 0.6830 | No | ||
| 19 | PITPNC1 | 7759 | 2713 | 0.152 | 0.6870 | No | ||
| 20 | EPHB2 | 4675 2440 8910 | 3107 | 0.095 | 0.6690 | No | ||
| 21 | KIF1B | 15671 2476 4950 15673 | 3421 | 0.065 | 0.6542 | No | ||
| 22 | HIPK1 | 4851 | 3567 | 0.055 | 0.6482 | No | ||
| 23 | ZNF385 | 22106 2297 | 3603 | 0.052 | 0.6481 | No | ||
| 24 | HOXB5 | 20687 | 3835 | 0.040 | 0.6370 | No | ||
| 25 | THRA | 1447 10171 1406 | 3856 | 0.039 | 0.6372 | No | ||
| 26 | IGF2R | 23123 | 4110 | 0.029 | 0.6245 | No | ||
| 27 | PBX1 | 5224 | 4148 | 0.028 | 0.6234 | No | ||
| 28 | LPHN1 | 6850 174 | 4337 | 0.023 | 0.6141 | No | ||
| 29 | EIF2C1 | 10672 | 4645 | 0.016 | 0.5980 | No | ||
| 30 | MAGI1 | 19828 4816 1179 | 4716 | 0.015 | 0.5948 | No | ||
| 31 | PCDHA5 | 8790 | 5096 | 0.011 | 0.5747 | No | ||
| 32 | CDC42 | 4503 8722 4504 2465 | 5208 | 0.010 | 0.5690 | No | ||
| 33 | RNF26 | 5614 | 5471 | 0.008 | 0.5552 | No | ||
| 34 | LUZP1 | 6533 11257 2503 | 5482 | 0.008 | 0.5549 | No | ||
| 35 | KLK5 | 1307 9237 | 5984 | 0.006 | 0.5281 | No | ||
| 36 | CORO2B | 6169 10652 | 6123 | 0.005 | 0.5208 | No | ||
| 37 | PRX | 3729 9628 | 6150 | 0.005 | 0.5196 | No | ||
| 38 | HSPA14 | 14695 | 6180 | 0.005 | 0.5182 | No | ||
| 39 | SLC30A3 | 5995 | 7184 | 0.002 | 0.4641 | No | ||
| 40 | CHES1 | 21007 11200 | 7323 | 0.002 | 0.4567 | No | ||
| 41 | PHYHIP | 21968 | 7808 | 0.000 | 0.4306 | No | ||
| 42 | SNTA1 | 14386 | 7919 | 0.000 | 0.4247 | No | ||
| 43 | SLC16A2 | 24086 | 8095 | -0.000 | 0.4153 | No | ||
| 44 | APBA1 | 11483 | 8426 | -0.001 | 0.3975 | No | ||
| 45 | PAK7 | 14417 | 9324 | -0.003 | 0.3492 | No | ||
| 46 | NEUROD2 | 20262 | 9400 | -0.003 | 0.3453 | No | ||
| 47 | TJP1 | 17803 | 9546 | -0.004 | 0.3376 | No | ||
| 48 | PCDHA12 | 9645 | 9560 | -0.004 | 0.3370 | No | ||
| 49 | KCNN3 | 15538 1913 | 9606 | -0.004 | 0.3347 | No | ||
| 50 | LRFN5 | 6213 | 9891 | -0.005 | 0.3195 | No | ||
| 51 | ADAMTS19 | 23544 | 10190 | -0.006 | 0.3036 | No | ||
| 52 | PCDHA3 | 9644 | 10342 | -0.006 | 0.2957 | No | ||
| 53 | BSN | 18997 | 10731 | -0.007 | 0.2750 | No | ||
| 54 | PCDHA10 | 8792 | 11291 | -0.009 | 0.2451 | No | ||
| 55 | PCDHA6 | 2038 8788 | 11312 | -0.009 | 0.2444 | No | ||
| 56 | BAI1 | 22459 | 11328 | -0.009 | 0.2439 | No | ||
| 57 | GAP43 | 9001 | 11393 | -0.010 | 0.2407 | No | ||
| 58 | NFASC | 11233 979 3952 4047 | 11722 | -0.011 | 0.2234 | No | ||
| 59 | ASB15 | 8129 | 11737 | -0.011 | 0.2230 | No | ||
| 60 | BRCA1 | 20213 | 11747 | -0.011 | 0.2229 | No | ||
| 61 | DMN | 6126 2542 17800 | 11809 | -0.012 | 0.2200 | No | ||
| 62 | PCDHA7 | 8789 | 12028 | -0.013 | 0.2087 | No | ||
| 63 | PAX2 | 9530 24416 | 12295 | -0.015 | 0.1948 | No | ||
| 64 | PCDHA1 | 8571 4368 | 12574 | -0.017 | 0.1804 | No | ||
| 65 | ASTN2 | 15860 2524 | 12678 | -0.018 | 0.1754 | No | ||
| 66 | NTRK3 | 2069 3652 3705 9492 | 12880 | -0.020 | 0.1652 | No | ||
| 67 | ST7L | 15465 1904 | 12906 | -0.020 | 0.1645 | No | ||
| 68 | RHOA | 8624 4409 4410 | 13271 | -0.024 | 0.1457 | No | ||
| 69 | FMO2 | 7063 | 13401 | -0.026 | 0.1396 | No | ||
| 70 | ZNF3 | 7094 3516 12097 | 13451 | -0.027 | 0.1378 | No | ||
| 71 | HCN4 | 9080 | 13468 | -0.027 | 0.1378 | No | ||
| 72 | HMGA2 | 4857 9098 3341 3444 | 13837 | -0.033 | 0.1191 | No | ||
| 73 | CPNE6 | 22010 | 13850 | -0.034 | 0.1196 | No | ||
| 74 | SLC37A2 | 19178 | 14091 | -0.039 | 0.1079 | No | ||
| 75 | PCDH10 | 5227 9534 1893 | 14116 | -0.039 | 0.1079 | No | ||
| 76 | ATP1A3 | 21 | 14557 | -0.053 | 0.0860 | No | ||
| 77 | PCDHA11 | 8791 | 15168 | -0.087 | 0.0559 | No | ||
| 78 | RHOT1 | 12227 | 15412 | -0.111 | 0.0465 | No | ||
| 79 | SNX19 | 3021 4165 | 16013 | -0.201 | 0.0209 | No | ||
| 80 | SERF2 | 11714 | 16017 | -0.201 | 0.0274 | No | ||
| 81 | ATP6V1F | 12291 | 16525 | -0.316 | 0.0106 | No | ||
| 82 | DCTN3 | 15908 | 17513 | -0.704 | -0.0192 | No | ||
| 83 | WARS | 2085 20984 | 17532 | -0.718 | 0.0038 | No | ||
| 84 | PRKAR2A | 5287 | 17536 | -0.722 | 0.0277 | No | ||
| 85 | RAE1 | 12395 | 17803 | -0.914 | 0.0439 | No |