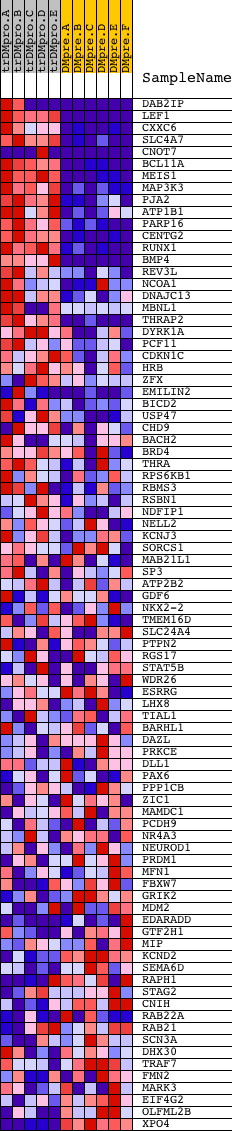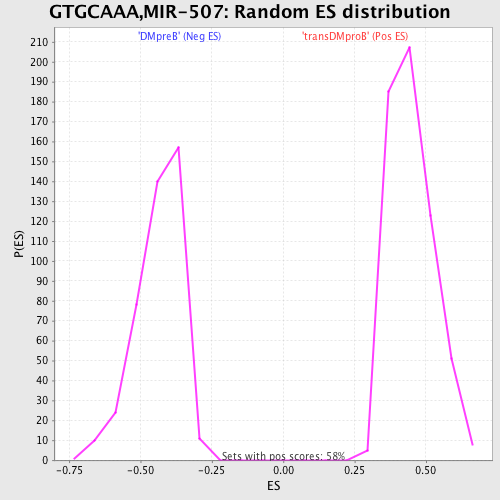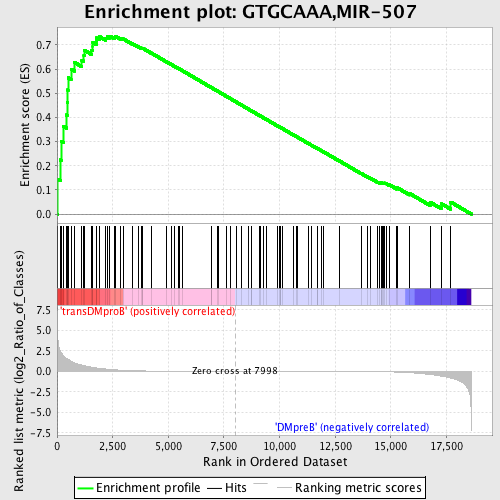
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | GTGCAAA,MIR-507 |
| Enrichment Score (ES) | 0.7366255 |
| Normalized Enrichment Score (NES) | 1.647943 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.021941386 |
| FWER p-Value | 0.085 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DAB2IP | 12808 2692 | 30 | 4.155 | 0.1452 | Yes | ||
| 2 | LEF1 | 1860 15420 | 146 | 2.413 | 0.2243 | Yes | ||
| 3 | CXXC6 | 19747 | 183 | 2.240 | 0.3015 | Yes | ||
| 4 | SLC4A7 | 10181 | 287 | 1.890 | 0.3627 | Yes | ||
| 5 | CNOT7 | 3787 9600 | 427 | 1.562 | 0.4104 | Yes | ||
| 6 | BCL11A | 4691 | 455 | 1.524 | 0.4628 | Yes | ||
| 7 | MEIS1 | 20524 | 481 | 1.480 | 0.5138 | Yes | ||
| 8 | MAP3K3 | 20626 | 496 | 1.448 | 0.5642 | Yes | ||
| 9 | PJA2 | 5904 | 631 | 1.222 | 0.6001 | Yes | ||
| 10 | ATP1B1 | 4420 | 786 | 1.009 | 0.6275 | Yes | ||
| 11 | PARP16 | 19405 | 1111 | 0.753 | 0.6366 | Yes | ||
| 12 | CENTG2 | 6874 | 1204 | 0.678 | 0.6556 | Yes | ||
| 13 | RUNX1 | 4481 | 1221 | 0.667 | 0.6783 | Yes | ||
| 14 | BMP4 | 21863 | 1540 | 0.507 | 0.6791 | Yes | ||
| 15 | REV3L | 20050 | 1602 | 0.479 | 0.6927 | Yes | ||
| 16 | NCOA1 | 5154 | 1605 | 0.478 | 0.7095 | Yes | ||
| 17 | DNAJC13 | 6177 | 1764 | 0.406 | 0.7153 | Yes | ||
| 18 | MBNL1 | 1921 15582 | 1771 | 0.402 | 0.7292 | Yes | ||
| 19 | THRAP2 | 8009 | 1900 | 0.356 | 0.7349 | Yes | ||
| 20 | DYRK1A | 4649 | 2191 | 0.269 | 0.7287 | Yes | ||
| 21 | PCF11 | 121 | 2247 | 0.256 | 0.7348 | Yes | ||
| 22 | CDKN1C | 17546 | 2365 | 0.230 | 0.7366 | Yes | ||
| 23 | HRB | 14208 4066 | 2573 | 0.186 | 0.7320 | No | ||
| 24 | ZFX | 5984 | 2613 | 0.175 | 0.7361 | No | ||
| 25 | EMILIN2 | 10901 | 2862 | 0.126 | 0.7272 | No | ||
| 26 | BICD2 | 8048 | 2975 | 0.108 | 0.7250 | No | ||
| 27 | USP47 | 6794 | 3394 | 0.067 | 0.7048 | No | ||
| 28 | CHD9 | 6784 11553 18531 | 3655 | 0.049 | 0.6925 | No | ||
| 29 | BACH2 | 8648 | 3777 | 0.043 | 0.6875 | No | ||
| 30 | BRD4 | 7164 1609 1628 1598 1552 1601 12176 1630 1501 | 3852 | 0.039 | 0.6849 | No | ||
| 31 | THRA | 1447 10171 1406 | 3856 | 0.039 | 0.6861 | No | ||
| 32 | RPS6KB1 | 7815 1207 13040 | 4229 | 0.026 | 0.6669 | No | ||
| 33 | RBMS3 | 18973 | 4896 | 0.013 | 0.6314 | No | ||
| 34 | RSBN1 | 6035 | 5132 | 0.011 | 0.6191 | No | ||
| 35 | NDFIP1 | 23573 | 5268 | 0.010 | 0.6122 | No | ||
| 36 | NELL2 | 12006 7020 22151 | 5461 | 0.008 | 0.6021 | No | ||
| 37 | KCNJ3 | 4944 | 5483 | 0.008 | 0.6013 | No | ||
| 38 | SORCS1 | 7184 12203 | 5620 | 0.007 | 0.5942 | No | ||
| 39 | MAB21L1 | 15589 5046 | 6938 | 0.003 | 0.5232 | No | ||
| 40 | SP3 | 5483 | 6954 | 0.003 | 0.5225 | No | ||
| 41 | ATP2B2 | 17038 | 7214 | 0.002 | 0.5086 | No | ||
| 42 | GDF6 | 10814 | 7234 | 0.002 | 0.5076 | No | ||
| 43 | NKX2-2 | 5176 | 7599 | 0.001 | 0.4880 | No | ||
| 44 | TMEM16D | 19649 | 7771 | 0.001 | 0.4788 | No | ||
| 45 | SLC24A4 | 6223 | 8067 | -0.000 | 0.4629 | No | ||
| 46 | PTPN2 | 23414 | 8291 | -0.001 | 0.4509 | No | ||
| 47 | RGS17 | 7133 | 8610 | -0.001 | 0.4338 | No | ||
| 48 | STAT5B | 20222 | 8729 | -0.002 | 0.4275 | No | ||
| 49 | WDR26 | 13733 | 8751 | -0.002 | 0.4264 | No | ||
| 50 | ESRRG | 6447 | 9116 | -0.003 | 0.4069 | No | ||
| 51 | LHX8 | 4997 | 9144 | -0.003 | 0.4055 | No | ||
| 52 | TIAL1 | 996 5753 | 9296 | -0.003 | 0.3975 | No | ||
| 53 | BARHL1 | 14634 | 9413 | -0.003 | 0.3913 | No | ||
| 54 | DAZL | 22944 | 9899 | -0.005 | 0.3653 | No | ||
| 55 | PRKCE | 9575 | 10001 | -0.005 | 0.3600 | No | ||
| 56 | DLL1 | 23119 | 10047 | -0.005 | 0.3578 | No | ||
| 57 | PAX6 | 5223 | 10062 | -0.005 | 0.3572 | No | ||
| 58 | PPP1CB | 5282 3529 3504 | 10127 | -0.005 | 0.3540 | No | ||
| 59 | ZIC1 | 19040 | 10643 | -0.007 | 0.3264 | No | ||
| 60 | MAMDC1 | 21054 6798 | 10774 | -0.007 | 0.3197 | No | ||
| 61 | PCDH9 | 21736 | 10825 | -0.007 | 0.3172 | No | ||
| 62 | NR4A3 | 9473 16212 5183 | 11303 | -0.009 | 0.2918 | No | ||
| 63 | NEUROD1 | 14550 | 11436 | -0.010 | 0.2850 | No | ||
| 64 | PRDM1 | 19775 3337 | 11687 | -0.011 | 0.2719 | No | ||
| 65 | MFN1 | 12529 1823 | 11717 | -0.011 | 0.2708 | No | ||
| 66 | FBXW7 | 1805 11928 | 11862 | -0.012 | 0.2634 | No | ||
| 67 | GRIK2 | 3318 | 11987 | -0.013 | 0.2572 | No | ||
| 68 | MDM2 | 19620 3327 | 12679 | -0.018 | 0.2205 | No | ||
| 69 | EDARADD | 5056 3282 3278 | 13681 | -0.030 | 0.1675 | No | ||
| 70 | GTF2H1 | 4069 18236 | 13954 | -0.036 | 0.1541 | No | ||
| 71 | MIP | 19845 | 14097 | -0.039 | 0.1478 | No | ||
| 72 | KCND2 | 4941 17515 | 14414 | -0.047 | 0.1324 | No | ||
| 73 | SEMA6D | 2800 14876 90 | 14510 | -0.051 | 0.1291 | No | ||
| 74 | RAPH1 | 19110 | 14567 | -0.053 | 0.1280 | No | ||
| 75 | STAG2 | 5521 | 14577 | -0.054 | 0.1294 | No | ||
| 76 | CNIH | 4536 23971 | 14642 | -0.057 | 0.1279 | No | ||
| 77 | RAB22A | 2888 5343 | 14687 | -0.059 | 0.1277 | No | ||
| 78 | RAB21 | 5696 | 14706 | -0.060 | 0.1288 | No | ||
| 79 | SCN3A | 14573 | 14783 | -0.063 | 0.1269 | No | ||
| 80 | DHX30 | 18989 | 14939 | -0.071 | 0.1211 | No | ||
| 81 | TRAF7 | 10325 | 15264 | -0.094 | 0.1069 | No | ||
| 82 | FMN2 | 14037 | 15297 | -0.098 | 0.1087 | No | ||
| 83 | MARK3 | 9369 | 15848 | -0.172 | 0.0851 | No | ||
| 84 | EIF4G2 | 1908 8892 | 16768 | -0.389 | 0.0492 | No | ||
| 85 | OLFML2B | 11496 | 17282 | -0.593 | 0.0425 | No | ||
| 86 | XPO4 | 7161 | 17690 | -0.833 | 0.0500 | No |