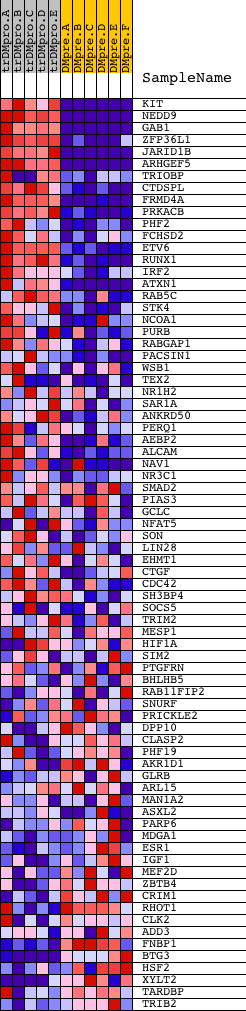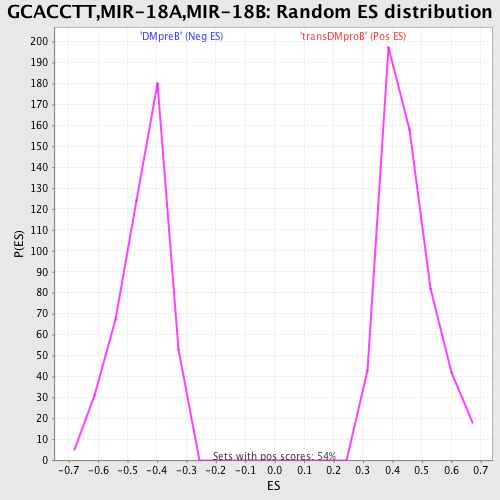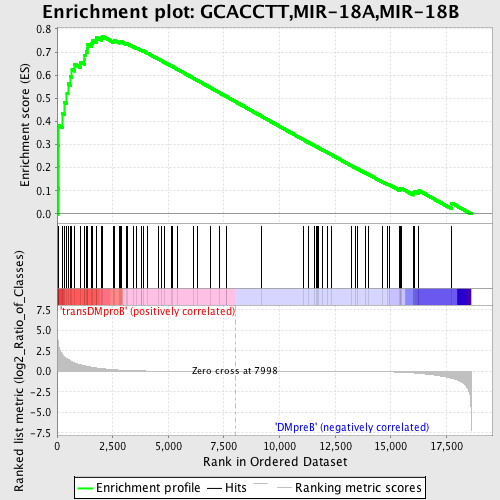
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | GCACCTT,MIR-18A,MIR-18B |
| Enrichment Score (ES) | 0.7687605 |
| Normalized Enrichment Score (NES) | 1.709156 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006958786 |
| FWER p-Value | 0.014 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KIT | 16823 | 40 | 3.755 | 0.1084 | Yes | ||
| 2 | NEDD9 | 3206 21483 | 61 | 3.267 | 0.2036 | Yes | ||
| 3 | GAB1 | 18828 | 64 | 3.185 | 0.2973 | Yes | ||
| 4 | ZFP36L1 | 4453 4454 | 83 | 2.920 | 0.3823 | Yes | ||
| 5 | JARID1B | 14123 | 241 | 2.037 | 0.4338 | Yes | ||
| 6 | ARHGEF5 | 12025 | 319 | 1.801 | 0.4827 | Yes | ||
| 7 | TRIOBP | 2280 670 8476 | 443 | 1.546 | 0.5216 | Yes | ||
| 8 | CTDSPL | 19279 | 490 | 1.460 | 0.5621 | Yes | ||
| 9 | FRMD4A | 5557 | 603 | 1.272 | 0.5936 | Yes | ||
| 10 | PRKACB | 15140 | 668 | 1.157 | 0.6242 | Yes | ||
| 11 | PHF2 | 21470 | 787 | 1.009 | 0.6475 | Yes | ||
| 12 | FCHSD2 | 18172 | 1060 | 0.793 | 0.6562 | Yes | ||
| 13 | ETV6 | 17264 | 1217 | 0.671 | 0.6676 | Yes | ||
| 14 | RUNX1 | 4481 | 1221 | 0.667 | 0.6870 | Yes | ||
| 15 | IRF2 | 18621 | 1298 | 0.623 | 0.7013 | Yes | ||
| 16 | ATXN1 | 21477 | 1349 | 0.598 | 0.7162 | Yes | ||
| 17 | RAB5C | 20226 | 1367 | 0.588 | 0.7326 | Yes | ||
| 18 | STK4 | 14743 | 1551 | 0.500 | 0.7375 | Yes | ||
| 19 | NCOA1 | 5154 | 1605 | 0.478 | 0.7487 | Yes | ||
| 20 | PURB | 5335 | 1757 | 0.410 | 0.7526 | Yes | ||
| 21 | RABGAP1 | 10439 | 1778 | 0.400 | 0.7633 | Yes | ||
| 22 | PACSIN1 | 6257 23322 10744 | 1995 | 0.324 | 0.7612 | Yes | ||
| 23 | WSB1 | 20330 | 2028 | 0.315 | 0.7688 | Yes | ||
| 24 | TEX2 | 5725 20181 20182 | 2542 | 0.192 | 0.7467 | No | ||
| 25 | NR1H2 | 17846 | 2581 | 0.183 | 0.7501 | No | ||
| 26 | SAR1A | 20008 | 2790 | 0.138 | 0.7429 | No | ||
| 27 | ANKRD50 | 533 | 2855 | 0.127 | 0.7432 | No | ||
| 28 | PERQ1 | 7168 | 2874 | 0.124 | 0.7459 | No | ||
| 29 | AEBP2 | 4359 | 3140 | 0.091 | 0.7343 | No | ||
| 30 | ALCAM | 4367 | 3162 | 0.088 | 0.7357 | No | ||
| 31 | NAV1 | 3978 5681 | 3448 | 0.063 | 0.7222 | No | ||
| 32 | NR3C1 | 9043 | 3588 | 0.053 | 0.7163 | No | ||
| 33 | SMAD2 | 23511 | 3778 | 0.043 | 0.7074 | No | ||
| 34 | PIAS3 | 15491 672 1906 | 3801 | 0.042 | 0.7074 | No | ||
| 35 | GCLC | 19374 | 3872 | 0.039 | 0.7048 | No | ||
| 36 | NFAT5 | 3921 7037 12036 | 4081 | 0.030 | 0.6944 | No | ||
| 37 | SON | 5473 1657 1684 | 4567 | 0.018 | 0.6688 | No | ||
| 38 | LIN28 | 15723 | 4680 | 0.016 | 0.6632 | No | ||
| 39 | EHMT1 | 13401 14670 2756 8090 | 4825 | 0.014 | 0.6559 | No | ||
| 40 | CTGF | 20068 | 5162 | 0.011 | 0.6380 | No | ||
| 41 | CDC42 | 4503 8722 4504 2465 | 5208 | 0.010 | 0.6359 | No | ||
| 42 | SH3BP4 | 8270 | 5424 | 0.009 | 0.6246 | No | ||
| 43 | SOCS5 | 1619 23141 | 6140 | 0.005 | 0.5862 | No | ||
| 44 | TRIM2 | 13485 8157 | 6310 | 0.005 | 0.5772 | No | ||
| 45 | MESP1 | 17786 | 6903 | 0.003 | 0.5453 | No | ||
| 46 | HIF1A | 4850 | 7307 | 0.002 | 0.5236 | No | ||
| 47 | SIM2 | 22693 | 7604 | 0.001 | 0.5077 | No | ||
| 48 | PTGFRN | 15224 | 7620 | 0.001 | 0.5069 | No | ||
| 49 | BHLHB5 | 15629 | 9205 | -0.003 | 0.4216 | No | ||
| 50 | RAB11FIP2 | 23635 | 11083 | -0.008 | 0.3206 | No | ||
| 51 | SNURF | 2251 | 11281 | -0.009 | 0.3102 | No | ||
| 52 | PRICKLE2 | 10837 | 11553 | -0.010 | 0.2959 | No | ||
| 53 | DPP10 | 13850 | 11670 | -0.011 | 0.2900 | No | ||
| 54 | CLASP2 | 13338 | 11682 | -0.011 | 0.2897 | No | ||
| 55 | PHF19 | 13150 | 11768 | -0.011 | 0.2855 | No | ||
| 56 | AKR1D1 | 17483 | 11944 | -0.012 | 0.2764 | No | ||
| 57 | GLRB | 15312 1846 | 12145 | -0.014 | 0.2660 | No | ||
| 58 | ARL15 | 21559 | 12335 | -0.015 | 0.2562 | No | ||
| 59 | MAN1A2 | 5076 5075 | 13218 | -0.024 | 0.2094 | No | ||
| 60 | ASXL2 | 7957 | 13413 | -0.026 | 0.1997 | No | ||
| 61 | PARP6 | 19419 2979 3156 | 13518 | -0.028 | 0.1949 | No | ||
| 62 | MDGA1 | 13231 | 13872 | -0.034 | 0.1768 | No | ||
| 63 | ESR1 | 20097 4685 | 13985 | -0.036 | 0.1719 | No | ||
| 64 | IGF1 | 3352 9156 3409 | 14645 | -0.057 | 0.1380 | No | ||
| 65 | MEF2D | 9379 | 14867 | -0.067 | 0.1281 | No | ||
| 66 | ZBTB4 | 13267 | 14952 | -0.072 | 0.1256 | No | ||
| 67 | CRIM1 | 403 | 15384 | -0.108 | 0.1056 | No | ||
| 68 | RHOT1 | 12227 | 15412 | -0.111 | 0.1074 | No | ||
| 69 | CLK2 | 1883 15544 | 15443 | -0.114 | 0.1091 | No | ||
| 70 | ADD3 | 23812 3694 | 15482 | -0.118 | 0.1106 | No | ||
| 71 | FNBP1 | 4732 | 16012 | -0.201 | 0.0879 | No | ||
| 72 | BTG3 | 8664 | 16014 | -0.201 | 0.0938 | No | ||
| 73 | HSF2 | 9128 | 16075 | -0.212 | 0.0968 | No | ||
| 74 | XYLT2 | 20288 | 16236 | -0.247 | 0.0955 | No | ||
| 75 | TARDBP | 10520 6066 | 16264 | -0.253 | 0.1015 | No | ||
| 76 | TRIB2 | 21102 | 17737 | -0.860 | 0.0474 | No |