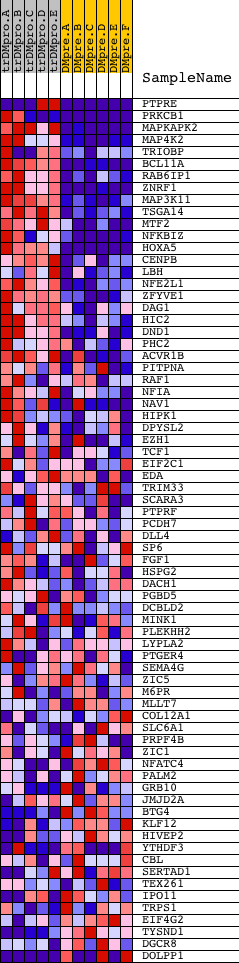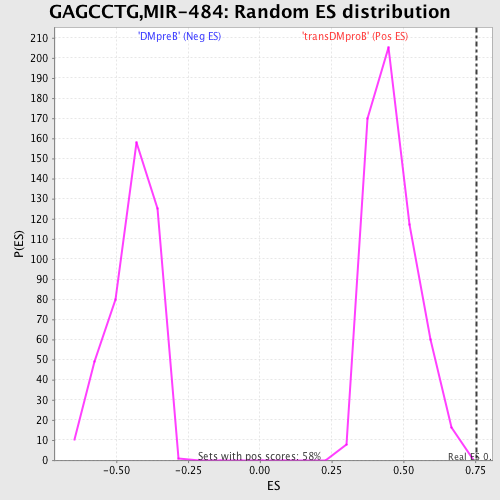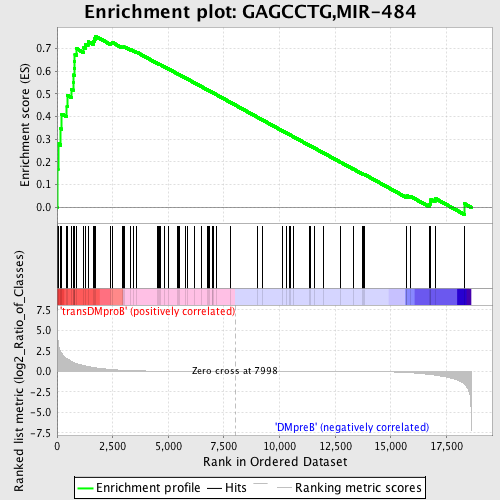
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | GAGCCTG,MIR-484 |
| Enrichment Score (ES) | 0.7539109 |
| Normalized Enrichment Score (NES) | 1.6402831 |
| Nominal p-value | 0.0017331023 |
| FDR q-value | 0.023658518 |
| FWER p-Value | 0.108 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTPRE | 1662 18039 | 7 | 5.433 | 0.1689 | Yes | ||
| 2 | PRKCB1 | 1693 9574 | 42 | 3.635 | 0.2803 | Yes | ||
| 3 | MAPKAPK2 | 13838 | 165 | 2.313 | 0.3458 | Yes | ||
| 4 | MAP4K2 | 6457 | 210 | 2.155 | 0.4106 | Yes | ||
| 5 | TRIOBP | 2280 670 8476 | 443 | 1.546 | 0.4462 | Yes | ||
| 6 | BCL11A | 4691 | 455 | 1.524 | 0.4931 | Yes | ||
| 7 | RAB6IP1 | 17676 | 641 | 1.206 | 0.5207 | Yes | ||
| 8 | ZNRF1 | 9306 | 725 | 1.080 | 0.5499 | Yes | ||
| 9 | MAP3K11 | 11163 | 734 | 1.072 | 0.5829 | Yes | ||
| 10 | TSGA14 | 17194 | 773 | 1.022 | 0.6127 | Yes | ||
| 11 | MTF2 | 5129 | 789 | 1.008 | 0.6433 | Yes | ||
| 12 | NFKBIZ | 22575 | 802 | 0.998 | 0.6738 | Yes | ||
| 13 | HOXA5 | 17149 | 880 | 0.923 | 0.6984 | Yes | ||
| 14 | CENPB | 14424 | 1199 | 0.681 | 0.7024 | Yes | ||
| 15 | LBH | 23160 | 1272 | 0.635 | 0.7183 | Yes | ||
| 16 | NFE2L1 | 9457 | 1392 | 0.578 | 0.7299 | Yes | ||
| 17 | ZFYVE1 | 10138 | 1637 | 0.463 | 0.7312 | Yes | ||
| 18 | DAG1 | 18996 8837 | 1692 | 0.439 | 0.7420 | Yes | ||
| 19 | HIC2 | 7185 | 1717 | 0.425 | 0.7539 | Yes | ||
| 20 | DND1 | 10017 | 2412 | 0.220 | 0.7233 | No | ||
| 21 | PHC2 | 16075 | 2487 | 0.203 | 0.7257 | No | ||
| 22 | ACVR1B | 4335 22353 | 2930 | 0.114 | 0.7054 | No | ||
| 23 | PITPNA | 20778 | 2976 | 0.108 | 0.7063 | No | ||
| 24 | RAF1 | 17035 | 3038 | 0.101 | 0.7062 | No | ||
| 25 | NFIA | 16172 5170 | 3302 | 0.075 | 0.6943 | No | ||
| 26 | NAV1 | 3978 5681 | 3448 | 0.063 | 0.6885 | No | ||
| 27 | HIPK1 | 4851 | 3567 | 0.055 | 0.6838 | No | ||
| 28 | DPYSL2 | 3122 4561 8787 | 4523 | 0.019 | 0.6329 | No | ||
| 29 | EZH1 | 20217 | 4561 | 0.018 | 0.6315 | No | ||
| 30 | TCF1 | 16416 | 4614 | 0.017 | 0.6292 | No | ||
| 31 | EIF2C1 | 10672 | 4645 | 0.016 | 0.6281 | No | ||
| 32 | EDA | 24284 | 4808 | 0.014 | 0.6198 | No | ||
| 33 | TRIM33 | 8236 13579 15472 8237 | 5009 | 0.012 | 0.6093 | No | ||
| 34 | SCARA3 | 21780 | 5403 | 0.009 | 0.5884 | No | ||
| 35 | PTPRF | 5331 2545 | 5451 | 0.008 | 0.5862 | No | ||
| 36 | PCDH7 | 7029 3460 | 5496 | 0.008 | 0.5840 | No | ||
| 37 | DLL4 | 7040 | 5789 | 0.007 | 0.5685 | No | ||
| 38 | SP6 | 8178 13678 13679 | 5838 | 0.006 | 0.5661 | No | ||
| 39 | FGF1 | 1994 23447 | 6176 | 0.005 | 0.5481 | No | ||
| 40 | HSPG2 | 4884 16023 | 6499 | 0.004 | 0.5309 | No | ||
| 41 | DACH1 | 4595 3371 | 6775 | 0.003 | 0.5161 | No | ||
| 42 | PGBD5 | 9960 | 6804 | 0.003 | 0.5147 | No | ||
| 43 | DCBLD2 | 7855 | 6839 | 0.003 | 0.5130 | No | ||
| 44 | MINK1 | 20802 | 6979 | 0.002 | 0.5055 | No | ||
| 45 | PLEKHH2 | 23148 | 6996 | 0.002 | 0.5048 | No | ||
| 46 | LYPLA2 | 15708 | 7041 | 0.002 | 0.5024 | No | ||
| 47 | PTGER4 | 9649 | 7171 | 0.002 | 0.4956 | No | ||
| 48 | SEMA4G | 11182 | 7790 | 0.000 | 0.4622 | No | ||
| 49 | ZIC5 | 12268 | 9023 | -0.002 | 0.3959 | No | ||
| 50 | M6PR | 1053 5044 9328 | 9240 | -0.003 | 0.3843 | No | ||
| 51 | MLLT7 | 24278 | 10144 | -0.005 | 0.3358 | No | ||
| 52 | COL12A1 | 3143 19054 | 10325 | -0.006 | 0.3263 | No | ||
| 53 | SLC6A1 | 10557 | 10446 | -0.006 | 0.3200 | No | ||
| 54 | PRPF4B | 3208 5295 | 10470 | -0.006 | 0.3190 | No | ||
| 55 | ZIC1 | 19040 | 10643 | -0.007 | 0.3099 | No | ||
| 56 | NFATC4 | 22002 | 11335 | -0.009 | 0.2729 | No | ||
| 57 | PALM2 | 10817 2368 | 11406 | -0.010 | 0.2695 | No | ||
| 58 | GRB10 | 4799 | 11584 | -0.010 | 0.2603 | No | ||
| 59 | JMJD2A | 10505 6058 | 11971 | -0.013 | 0.2398 | No | ||
| 60 | BTG4 | 19456 | 12744 | -0.018 | 0.1988 | No | ||
| 61 | KLF12 | 4960 2637 21732 9228 | 13305 | -0.025 | 0.1693 | No | ||
| 62 | HIVEP2 | 9091 | 13748 | -0.032 | 0.1465 | No | ||
| 63 | YTHDF3 | 15630 10469 6019 | 13778 | -0.032 | 0.1459 | No | ||
| 64 | CBL | 19154 | 13815 | -0.033 | 0.1450 | No | ||
| 65 | SERTAD1 | 18326 | 15711 | -0.149 | 0.0475 | No | ||
| 66 | TEX261 | 17094 5726 | 15725 | -0.151 | 0.0515 | No | ||
| 67 | IPO11 | 21353 | 15867 | -0.175 | 0.0493 | No | ||
| 68 | TRPS1 | 8195 | 16727 | -0.376 | 0.0147 | No | ||
| 69 | EIF4G2 | 1908 8892 | 16768 | -0.389 | 0.0247 | No | ||
| 70 | TYSND1 | 20007 | 16800 | -0.397 | 0.0354 | No | ||
| 71 | DGCR8 | 22644 | 16990 | -0.469 | 0.0398 | No | ||
| 72 | DOLPP1 | 2682 15051 | 18307 | -1.535 | 0.0167 | No |