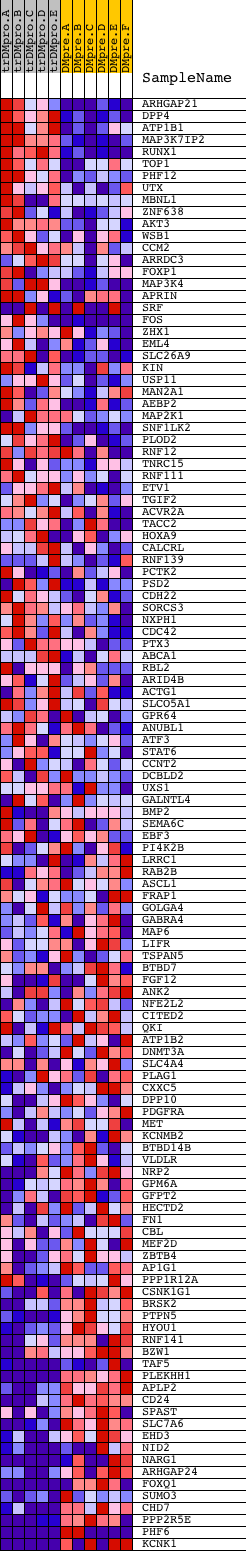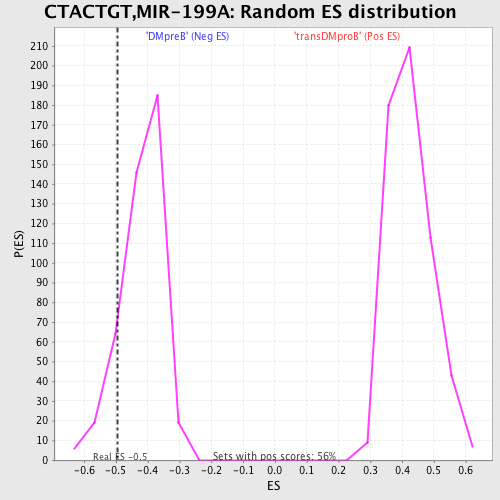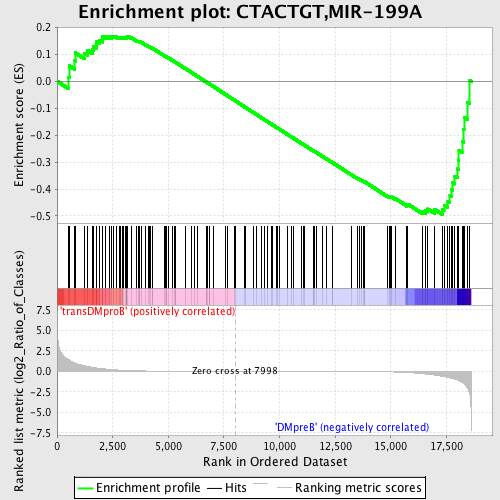
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | CTACTGT,MIR-199A |
| Enrichment Score (ES) | -0.4945555 |
| Normalized Enrichment Score (NES) | -1.1859521 |
| Nominal p-value | 0.12984055 |
| FDR q-value | 0.7722451 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARHGAP21 | 12929 | 527 | 1.406 | 0.0156 | No | ||
| 2 | DPP4 | 14579 | 544 | 1.368 | 0.0576 | No | ||
| 3 | ATP1B1 | 4420 | 786 | 1.009 | 0.0762 | No | ||
| 4 | MAP3K7IP2 | 19827 | 816 | 0.979 | 0.1053 | No | ||
| 5 | RUNX1 | 4481 | 1221 | 0.667 | 0.1043 | No | ||
| 6 | TOP1 | 5790 5789 10210 | 1379 | 0.582 | 0.1141 | No | ||
| 7 | PHF12 | 11192 6483 | 1577 | 0.490 | 0.1188 | No | ||
| 8 | UTX | 10266 2574 | 1640 | 0.462 | 0.1299 | No | ||
| 9 | MBNL1 | 1921 15582 | 1771 | 0.402 | 0.1355 | No | ||
| 10 | ZNF638 | 9476 | 1785 | 0.397 | 0.1472 | No | ||
| 11 | AKT3 | 13739 982 | 1896 | 0.357 | 0.1525 | No | ||
| 12 | WSB1 | 20330 | 2028 | 0.315 | 0.1552 | No | ||
| 13 | CCM2 | 20950 | 2040 | 0.312 | 0.1644 | No | ||
| 14 | ARRDC3 | 4191 | 2168 | 0.274 | 0.1662 | No | ||
| 15 | FOXP1 | 4242 | 2341 | 0.235 | 0.1642 | No | ||
| 16 | MAP3K4 | 23126 | 2446 | 0.212 | 0.1653 | No | ||
| 17 | APRIN | 4145 | 2526 | 0.195 | 0.1671 | No | ||
| 18 | SRF | 22961 1597 | 2654 | 0.165 | 0.1654 | No | ||
| 19 | FOS | 21202 | 2816 | 0.135 | 0.1609 | No | ||
| 20 | ZHX1 | 10428 5990 | 2866 | 0.125 | 0.1622 | No | ||
| 21 | EML4 | 8122 | 2937 | 0.113 | 0.1620 | No | ||
| 22 | SLC26A9 | 11560 980 4067 | 2986 | 0.107 | 0.1627 | No | ||
| 23 | KIN | 15120 | 3069 | 0.098 | 0.1614 | No | ||
| 24 | USP11 | 24368 10678 | 3115 | 0.094 | 0.1619 | No | ||
| 25 | MAN2A1 | 23170 | 3130 | 0.092 | 0.1640 | No | ||
| 26 | AEBP2 | 4359 | 3140 | 0.091 | 0.1664 | No | ||
| 27 | MAP2K1 | 19082 | 3174 | 0.087 | 0.1673 | No | ||
| 28 | SNF1LK2 | 6168 10649 | 3322 | 0.073 | 0.1617 | No | ||
| 29 | PLOD2 | 19352 | 3576 | 0.054 | 0.1497 | No | ||
| 30 | RNF12 | 5384 | 3639 | 0.050 | 0.1479 | No | ||
| 31 | TNRC15 | 5971 | 3691 | 0.047 | 0.1466 | No | ||
| 32 | RNF111 | 13555 8218 13556 | 3710 | 0.046 | 0.1471 | No | ||
| 33 | ETV1 | 4688 8925 8924 | 3791 | 0.042 | 0.1441 | No | ||
| 34 | TGIF2 | 6013 6012 | 3986 | 0.033 | 0.1346 | No | ||
| 35 | ACVR2A | 8542 | 4102 | 0.029 | 0.1293 | No | ||
| 36 | TACC2 | 18052 2534 2234 2435 2350 | 4142 | 0.028 | 0.1281 | No | ||
| 37 | HOXA9 | 17147 9109 1024 | 4211 | 0.026 | 0.1253 | No | ||
| 38 | CALCRL | 12041 | 4265 | 0.025 | 0.1232 | No | ||
| 39 | RNF139 | 7992 13291 | 4297 | 0.024 | 0.1223 | No | ||
| 40 | PCTK2 | 19907 | 4830 | 0.014 | 0.0939 | No | ||
| 41 | PSD2 | 23597 | 4879 | 0.013 | 0.0917 | No | ||
| 42 | CDH22 | 14340 | 4918 | 0.013 | 0.0901 | No | ||
| 43 | SORCS3 | 7323 | 4928 | 0.013 | 0.0900 | No | ||
| 44 | NXPH1 | 17524 | 4998 | 0.012 | 0.0866 | No | ||
| 45 | CDC42 | 4503 8722 4504 2465 | 5208 | 0.010 | 0.0757 | No | ||
| 46 | PTX3 | 9668 15575 | 5257 | 0.010 | 0.0734 | No | ||
| 47 | ABCA1 | 15881 | 5304 | 0.010 | 0.0712 | No | ||
| 48 | RBL2 | 18530 3781 | 5332 | 0.009 | 0.0700 | No | ||
| 49 | ARID4B | 21705 3233 | 5765 | 0.007 | 0.0469 | No | ||
| 50 | ACTG1 | 8535 | 6034 | 0.006 | 0.0326 | No | ||
| 51 | SLCO5A1 | 6286 | 6173 | 0.005 | 0.0253 | No | ||
| 52 | GPR64 | 24217 2644 | 6287 | 0.005 | 0.0193 | No | ||
| 53 | ANUBL1 | 17312 | 6702 | 0.003 | -0.0030 | No | ||
| 54 | ATF3 | 13720 | 6713 | 0.003 | -0.0034 | No | ||
| 55 | STAT6 | 19854 9909 | 6764 | 0.003 | -0.0060 | No | ||
| 56 | CCNT2 | 3993 13079 | 6835 | 0.003 | -0.0097 | No | ||
| 57 | DCBLD2 | 7855 | 6839 | 0.003 | -0.0098 | No | ||
| 58 | UXS1 | 13965 | 7012 | 0.002 | -0.0190 | No | ||
| 59 | GALNTL4 | 10599 | 7589 | 0.001 | -0.0501 | No | ||
| 60 | BMP2 | 14833 | 7661 | 0.001 | -0.0540 | No | ||
| 61 | SEMA6C | 1797 5424 | 7952 | 0.000 | -0.0696 | No | ||
| 62 | EBF3 | 8878 | 8011 | -0.000 | -0.0728 | No | ||
| 63 | PI4K2B | 3511 16847 | 8416 | -0.001 | -0.0946 | No | ||
| 64 | LRRC1 | 19059 3129 | 8489 | -0.001 | -0.0984 | No | ||
| 65 | RAB2B | 21842 | 8842 | -0.002 | -0.1174 | No | ||
| 66 | ASCL1 | 19655 | 8982 | -0.002 | -0.1248 | No | ||
| 67 | FRAP1 | 2468 15991 | 9178 | -0.003 | -0.1353 | No | ||
| 68 | GOLGA4 | 12022 | 9317 | -0.003 | -0.1427 | No | ||
| 69 | GABRA4 | 16519 | 9444 | -0.003 | -0.1494 | No | ||
| 70 | MAP6 | 9419 | 9619 | -0.004 | -0.1586 | No | ||
| 71 | LIFR | 22515 9277 | 9698 | -0.004 | -0.1627 | No | ||
| 72 | TSPAN5 | 15402 | 9878 | -0.005 | -0.1723 | No | ||
| 73 | BTBD7 | 20998 | 9915 | -0.005 | -0.1741 | No | ||
| 74 | FGF12 | 1723 22621 | 9991 | -0.005 | -0.1780 | No | ||
| 75 | ANK2 | 4275 | 10350 | -0.006 | -0.1971 | No | ||
| 76 | NFE2L2 | 2898 14557 | 10545 | -0.007 | -0.2074 | No | ||
| 77 | CITED2 | 5118 14477 | 10610 | -0.007 | -0.2107 | No | ||
| 78 | QKI | 23128 5340 | 10970 | -0.008 | -0.2298 | No | ||
| 79 | ATP1B2 | 20390 | 11084 | -0.008 | -0.2357 | No | ||
| 80 | DNMT3A | 2167 21330 | 11103 | -0.008 | -0.2364 | No | ||
| 81 | SLC4A4 | 16807 12035 7036 | 11521 | -0.010 | -0.2586 | No | ||
| 82 | PLAG1 | 7148 15949 12161 | 11523 | -0.010 | -0.2583 | No | ||
| 83 | CXXC5 | 12523 | 11551 | -0.010 | -0.2595 | No | ||
| 84 | DPP10 | 13850 | 11670 | -0.011 | -0.2655 | No | ||
| 85 | PDGFRA | 16824 | 11924 | -0.012 | -0.2788 | No | ||
| 86 | MET | 17520 | 12124 | -0.013 | -0.2891 | No | ||
| 87 | KCNMB2 | 1939 15615 | 12376 | -0.015 | -0.3022 | No | ||
| 88 | BTBD14B | 7344 7343 | 12391 | -0.015 | -0.3025 | No | ||
| 89 | VLDLR | 5858 3763 912 3682 | 13213 | -0.023 | -0.3462 | No | ||
| 90 | NRP2 | 5194 9486 5195 | 13510 | -0.028 | -0.3613 | No | ||
| 91 | GPM6A | 3834 10618 | 13586 | -0.029 | -0.3644 | No | ||
| 92 | GFPT2 | 4775 | 13664 | -0.030 | -0.3677 | No | ||
| 93 | HECTD2 | 23876 3721 | 13760 | -0.032 | -0.3718 | No | ||
| 94 | FN1 | 4091 4094 971 13929 | 13809 | -0.033 | -0.3734 | No | ||
| 95 | CBL | 19154 | 13815 | -0.033 | -0.3726 | No | ||
| 96 | MEF2D | 9379 | 14867 | -0.067 | -0.4273 | No | ||
| 97 | ZBTB4 | 13267 | 14952 | -0.072 | -0.4296 | No | ||
| 98 | AP1G1 | 4392 | 14995 | -0.074 | -0.4296 | No | ||
| 99 | PPP1R12A | 3354 19886 | 15021 | -0.076 | -0.4285 | No | ||
| 100 | CSNK1G1 | 3057 5671 19397 | 15205 | -0.090 | -0.4356 | No | ||
| 101 | BRSK2 | 190 | 15722 | -0.150 | -0.4588 | No | ||
| 102 | PTPN5 | 17817 | 15755 | -0.154 | -0.4557 | No | ||
| 103 | HYOU1 | 19476 | 16441 | -0.294 | -0.4835 | Yes | ||
| 104 | RNF141 | 12478 7392 | 16567 | -0.329 | -0.4800 | Yes | ||
| 105 | BZW1 | 7356 | 16668 | -0.360 | -0.4741 | Yes | ||
| 106 | TAF5 | 23833 5934 3759 | 16975 | -0.465 | -0.4761 | Yes | ||
| 107 | PLEKHH1 | 21229 | 17318 | -0.607 | -0.4756 | Yes | ||
| 108 | APLP2 | 19190 | 17394 | -0.639 | -0.4596 | Yes | ||
| 109 | CD24 | 8711 | 17564 | -0.741 | -0.4455 | Yes | ||
| 110 | SPAST | 1607 23158 11938 6943 | 17657 | -0.807 | -0.4252 | Yes | ||
| 111 | SLC7A6 | 18483 | 17735 | -0.859 | -0.4024 | Yes | ||
| 112 | EHD3 | 23159 | 17776 | -0.898 | -0.3765 | Yes | ||
| 113 | NID2 | 22089 | 17870 | -0.945 | -0.3519 | Yes | ||
| 114 | NARG1 | 7934 917 | 17996 | -1.066 | -0.3252 | Yes | ||
| 115 | ARHGAP24 | 16778 3655 | 18037 | -1.119 | -0.2923 | Yes | ||
| 116 | FOXQ1 | 21678 | 18062 | -1.152 | -0.2575 | Yes | ||
| 117 | SUMO3 | 5464 9837 | 18204 | -1.357 | -0.2226 | Yes | ||
| 118 | CHD7 | 16278 | 18282 | -1.479 | -0.1805 | Yes | ||
| 119 | PPP2R5E | 6523 2136 | 18291 | -1.501 | -0.1339 | Yes | ||
| 120 | PHF6 | 2648 24340 | 18452 | -2.076 | -0.0775 | Yes | ||
| 121 | KCNK1 | 18415 | 18550 | -2.754 | 0.0036 | Yes |