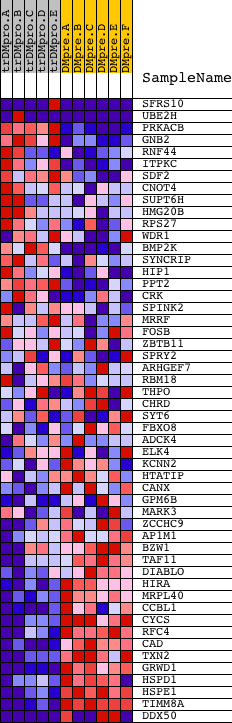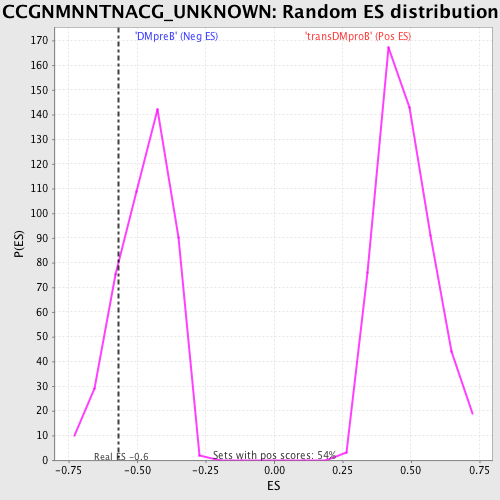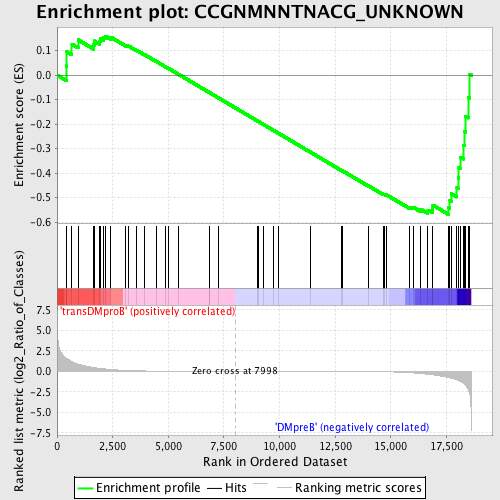
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | CCGNMNNTNACG_UNKNOWN |
| Enrichment Score (ES) | -0.5685483 |
| Normalized Enrichment Score (NES) | -1.2024693 |
| Nominal p-value | 0.17286652 |
| FDR q-value | 0.81469834 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SFRS10 | 9818 1722 5441 | 413 | 1.582 | 0.0373 | No | ||
| 2 | UBE2H | 5823 5822 | 429 | 1.561 | 0.0953 | No | ||
| 3 | PRKACB | 15140 | 668 | 1.157 | 0.1260 | No | ||
| 4 | GNB2 | 9026 | 947 | 0.868 | 0.1437 | No | ||
| 5 | RNF44 | 8364 | 1643 | 0.461 | 0.1236 | No | ||
| 6 | ITPKC | 17919 | 1667 | 0.451 | 0.1393 | No | ||
| 7 | SDF2 | 20762 | 1919 | 0.349 | 0.1390 | No | ||
| 8 | CNOT4 | 7010 1022 | 1958 | 0.336 | 0.1496 | No | ||
| 9 | SUPT6H | 9940 | 2100 | 0.293 | 0.1530 | No | ||
| 10 | HMG20B | 19676 | 2179 | 0.273 | 0.1591 | No | ||
| 11 | RPS27 | 12183 | 2407 | 0.220 | 0.1551 | No | ||
| 12 | WDR1 | 16544 3592 | 3092 | 0.096 | 0.1219 | No | ||
| 13 | BMP2K | 3657 16786 | 3208 | 0.084 | 0.1188 | No | ||
| 14 | SYNCRIP | 3078 3035 3107 | 3575 | 0.054 | 0.1012 | No | ||
| 15 | HIP1 | 5676 | 3935 | 0.036 | 0.0832 | No | ||
| 16 | PPT2 | 1617 12034 | 4476 | 0.020 | 0.0548 | No | ||
| 17 | CRK | 4559 1249 | 4892 | 0.013 | 0.0330 | No | ||
| 18 | SPINK2 | 16502 | 5024 | 0.012 | 0.0263 | No | ||
| 19 | MRRF | 12594 2837 | 5476 | 0.008 | 0.0024 | No | ||
| 20 | FOSB | 17945 | 6837 | 0.003 | -0.0708 | No | ||
| 21 | ZBTB11 | 11309 | 6856 | 0.003 | -0.0717 | No | ||
| 22 | SPRY2 | 21725 | 7275 | 0.002 | -0.0941 | No | ||
| 23 | ARHGEF7 | 3823 | 8994 | -0.002 | -0.1866 | No | ||
| 24 | RBM18 | 2789 14604 | 9066 | -0.003 | -0.1903 | No | ||
| 25 | THPO | 22636 | 9280 | -0.003 | -0.2016 | No | ||
| 26 | CHRD | 22817 | 9734 | -0.004 | -0.2259 | No | ||
| 27 | SYT6 | 13437 7042 12040 | 9928 | -0.005 | -0.2361 | No | ||
| 28 | FBXO8 | 18616 3896 | 11367 | -0.009 | -0.3132 | No | ||
| 29 | ADCK4 | 13364 | 12796 | -0.019 | -0.3894 | No | ||
| 30 | ELK4 | 8895 4664 3937 | 12836 | -0.019 | -0.3908 | No | ||
| 31 | KCNN2 | 8933 1949 | 13975 | -0.036 | -0.4507 | No | ||
| 32 | HTATIP | 3690 | 14680 | -0.059 | -0.4864 | No | ||
| 33 | CANX | 4474 | 14714 | -0.060 | -0.4859 | No | ||
| 34 | GPM6B | 9034 | 14812 | -0.064 | -0.4887 | No | ||
| 35 | MARK3 | 9369 | 15848 | -0.172 | -0.5380 | No | ||
| 36 | ZCCHC9 | 21398 | 16000 | -0.199 | -0.5387 | No | ||
| 37 | AP1M1 | 18571 | 16333 | -0.269 | -0.5465 | No | ||
| 38 | BZW1 | 7356 | 16668 | -0.360 | -0.5509 | Yes | ||
| 39 | TAF11 | 12733 | 16877 | -0.426 | -0.5461 | Yes | ||
| 40 | DIABLO | 16375 | 16890 | -0.433 | -0.5304 | Yes | ||
| 41 | HIRA | 4852 9090 | 17599 | -0.761 | -0.5399 | Yes | ||
| 42 | MRPL40 | 22641 | 17655 | -0.805 | -0.5126 | Yes | ||
| 43 | CCBL1 | 14630 | 17732 | -0.857 | -0.4844 | Yes | ||
| 44 | CYCS | 8821 | 17971 | -1.039 | -0.4581 | Yes | ||
| 45 | RFC4 | 1735 22627 | 18026 | -1.106 | -0.4194 | Yes | ||
| 46 | CAD | 16886 | 18060 | -1.150 | -0.3779 | Yes | ||
| 47 | TXN2 | 22227 | 18138 | -1.257 | -0.3347 | Yes | ||
| 48 | GRWD1 | 8317 | 18280 | -1.471 | -0.2870 | Yes | ||
| 49 | HSPD1 | 4078 9129 | 18318 | -1.562 | -0.2302 | Yes | ||
| 50 | HSPE1 | 9133 | 18364 | -1.702 | -0.1686 | Yes | ||
| 51 | TIMM8A | 24062 | 18484 | -2.233 | -0.0909 | Yes | ||
| 52 | DDX50 | 3324 19749 | 18539 | -2.603 | 0.0041 | Yes |