Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
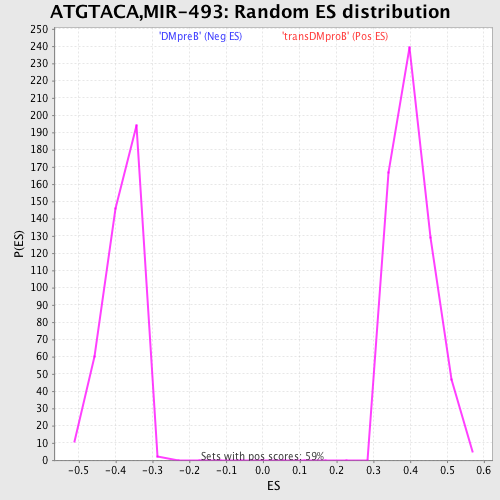
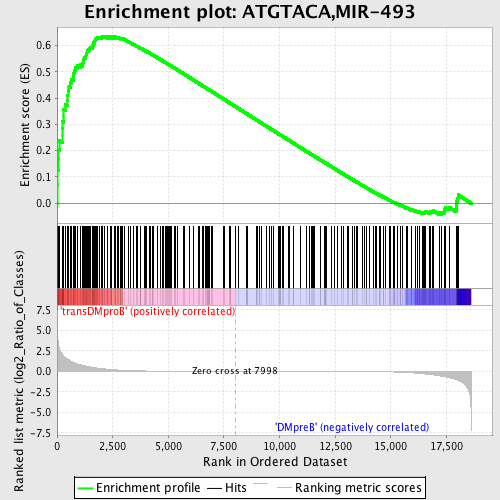
| GeneSet | ATGTACA,MIR-493 |
| Enrichment Score (ES) | 0.6355392 |
| Normalized Enrichment Score (NES) | 1.5746979 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.043675985 |
| FWER p-Value | 0.377 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 6 | 5.440 | 0.0716 | Yes | ||
| 2 | DAB2IP | 12808 2692 | 30 | 4.155 | 0.1253 | Yes | ||
| 3 | GAB1 | 18828 | 64 | 3.185 | 0.1656 | Yes | ||
| 4 | KLF3 | 4961 | 82 | 2.952 | 0.2037 | Yes | ||
| 5 | MAPK1 | 1642 11167 | 108 | 2.667 | 0.2376 | Yes | ||
| 6 | DOCK9 | 8369 | 240 | 2.049 | 0.2576 | Yes | ||
| 7 | JARID1B | 14123 | 241 | 2.037 | 0.2845 | Yes | ||
| 8 | CUGBP2 | 4687 8923 | 251 | 2.014 | 0.3106 | Yes | ||
| 9 | FOXO1A | 7124 | 297 | 1.842 | 0.3325 | Yes | ||
| 10 | RBM12 | 7980 2710 13280 | 299 | 1.837 | 0.3568 | Yes | ||
| 11 | FMNL2 | 15012 | 373 | 1.657 | 0.3747 | Yes | ||
| 12 | FGF13 | 4720 8963 2627 | 450 | 1.532 | 0.3908 | Yes | ||
| 13 | AXIN1 | 1579 23330 | 461 | 1.518 | 0.4104 | Yes | ||
| 14 | CTDSPL | 19279 | 490 | 1.460 | 0.4281 | Yes | ||
| 15 | MBNL2 | 21932 | 532 | 1.403 | 0.4444 | Yes | ||
| 16 | VGLL4 | 10558 | 602 | 1.273 | 0.4575 | Yes | ||
| 17 | BCL7A | 8063 | 628 | 1.227 | 0.4724 | Yes | ||
| 18 | CREBBP | 22682 8783 | 740 | 1.067 | 0.4805 | Yes | ||
| 19 | HMGCS1 | 9917 | 749 | 1.050 | 0.4939 | Yes | ||
| 20 | PHF2 | 21470 | 787 | 1.009 | 0.5052 | Yes | ||
| 21 | SP4 | 20972 | 820 | 0.976 | 0.5164 | Yes | ||
| 22 | PDPK1 | 23097 | 914 | 0.896 | 0.5232 | Yes | ||
| 23 | CHKB | 4518 | 1036 | 0.810 | 0.5273 | Yes | ||
| 24 | BAZ2A | 4371 | 1139 | 0.727 | 0.5314 | Yes | ||
| 25 | MCL1 | 15502 | 1166 | 0.703 | 0.5393 | Yes | ||
| 26 | CDC14A | 15184 | 1168 | 0.702 | 0.5485 | Yes | ||
| 27 | TBK1 | 12137 | 1211 | 0.674 | 0.5551 | Yes | ||
| 28 | MEF2C | 3204 9378 | 1283 | 0.629 | 0.5596 | Yes | ||
| 29 | IRF2 | 18621 | 1298 | 0.623 | 0.5670 | Yes | ||
| 30 | UBE2G1 | 12472 7390 1353 | 1342 | 0.601 | 0.5727 | Yes | ||
| 31 | LRP12 | 22304 | 1344 | 0.600 | 0.5805 | Yes | ||
| 32 | HNRPA0 | 13382 | 1411 | 0.566 | 0.5844 | Yes | ||
| 33 | VAMP2 | 20826 | 1445 | 0.551 | 0.5899 | Yes | ||
| 34 | ITSN2 | 21327 | 1508 | 0.523 | 0.5935 | Yes | ||
| 35 | PHF12 | 11192 6483 | 1577 | 0.490 | 0.5963 | Yes | ||
| 36 | NCOA1 | 5154 | 1605 | 0.478 | 0.6011 | Yes | ||
| 37 | PIAS1 | 7126 | 1633 | 0.464 | 0.6058 | Yes | ||
| 38 | UTX | 10266 2574 | 1640 | 0.462 | 0.6116 | Yes | ||
| 39 | SPEN | 12112 | 1688 | 0.440 | 0.6148 | Yes | ||
| 40 | FBS1 | 18067 | 1702 | 0.432 | 0.6198 | Yes | ||
| 41 | HIC2 | 7185 | 1717 | 0.425 | 0.6247 | Yes | ||
| 42 | DNAJC13 | 6177 | 1764 | 0.406 | 0.6275 | Yes | ||
| 43 | PPP3CB | 5285 | 1794 | 0.394 | 0.6312 | Yes | ||
| 44 | THRAP2 | 8009 | 1900 | 0.356 | 0.6302 | Yes | ||
| 45 | IRF7 | 12008 | 1991 | 0.326 | 0.6296 | Yes | ||
| 46 | RBBP6 | 5365 | 2017 | 0.318 | 0.6324 | Yes | ||
| 47 | JUN | 15832 | 2037 | 0.313 | 0.6355 | Yes | ||
| 48 | RYBP | 17056 | 2115 | 0.290 | 0.6352 | No | ||
| 49 | GNB1 | 15967 | 2253 | 0.254 | 0.6311 | No | ||
| 50 | BRD7 | 6540 3833 | 2285 | 0.247 | 0.6327 | No | ||
| 51 | CLASP1 | 14159 | 2392 | 0.223 | 0.6299 | No | ||
| 52 | USP21 | 13762 | 2452 | 0.211 | 0.6295 | No | ||
| 53 | AP3S1 | 8604 | 2457 | 0.210 | 0.6320 | No | ||
| 54 | KBTBD2 | 17137 | 2461 | 0.209 | 0.6346 | No | ||
| 55 | PPP1R10 | 6972 11960 6971 | 2593 | 0.179 | 0.6299 | No | ||
| 56 | SH2D3C | 2864 | 2605 | 0.176 | 0.6316 | No | ||
| 57 | ZFX | 5984 | 2613 | 0.175 | 0.6335 | No | ||
| 58 | SIPA1L2 | 10863 | 2715 | 0.151 | 0.6300 | No | ||
| 59 | TNFSF11 | 21745 | 2756 | 0.144 | 0.6298 | No | ||
| 60 | ANKRD50 | 533 | 2855 | 0.127 | 0.6261 | No | ||
| 61 | SOX21 | 27 10278 2778 | 2857 | 0.127 | 0.6277 | No | ||
| 62 | HEMGN | 15888 | 2897 | 0.119 | 0.6272 | No | ||
| 63 | ITGB1 | 3872 18411 | 2943 | 0.113 | 0.6262 | No | ||
| 64 | MAST1 | 12143 | 3019 | 0.103 | 0.6235 | No | ||
| 65 | PBX3 | 9531 2714 2705 | 3215 | 0.083 | 0.6140 | No | ||
| 66 | SS18 | 6506 6505 2030 | 3301 | 0.076 | 0.6104 | No | ||
| 67 | KIF1B | 15671 2476 4950 15673 | 3421 | 0.065 | 0.6048 | No | ||
| 68 | HIPK1 | 4851 | 3567 | 0.055 | 0.5976 | No | ||
| 69 | THRAP1 | 20309 | 3592 | 0.053 | 0.5970 | No | ||
| 70 | GTF2IRD1 | 7159 3635 3600 | 3735 | 0.045 | 0.5899 | No | ||
| 71 | SSH2 | 6202 | 3751 | 0.044 | 0.5897 | No | ||
| 72 | MLLT10 | 9393 5104 | 3759 | 0.044 | 0.5899 | No | ||
| 73 | MADD | 2926 14526 | 3906 | 0.037 | 0.5824 | No | ||
| 74 | EYA1 | 4695 4061 | 3951 | 0.035 | 0.5805 | No | ||
| 75 | AFF3 | 4982 | 3983 | 0.033 | 0.5792 | No | ||
| 76 | TTC7B | 21006 | 4021 | 0.032 | 0.5777 | No | ||
| 77 | PDE4D | 10722 6235 | 4163 | 0.028 | 0.5704 | No | ||
| 78 | SVIL | 1990 1942 10334 1976 23626 1946 1945 2041 | 4208 | 0.026 | 0.5683 | No | ||
| 79 | RNF38 | 7862 2328 13115 | 4276 | 0.025 | 0.5650 | No | ||
| 80 | BCOR | 12934 2561 | 4302 | 0.024 | 0.5640 | No | ||
| 81 | SV2B | 17795 | 4340 | 0.023 | 0.5622 | No | ||
| 82 | NAB1 | 9442 9443 5145 5144 | 4525 | 0.018 | 0.5525 | No | ||
| 83 | EIF2C1 | 10672 | 4645 | 0.016 | 0.5462 | No | ||
| 84 | ANKRD17 | 13496 3474 8170 | 4750 | 0.015 | 0.5408 | No | ||
| 85 | CSNK1A1 | 8204 | 4783 | 0.014 | 0.5392 | No | ||
| 86 | PSD2 | 23597 | 4879 | 0.013 | 0.5342 | No | ||
| 87 | SORCS3 | 7323 | 4928 | 0.013 | 0.5318 | No | ||
| 88 | PIK3R1 | 3170 | 4940 | 0.013 | 0.5314 | No | ||
| 89 | NXPH1 | 17524 | 4998 | 0.012 | 0.5284 | No | ||
| 90 | PMP22 | 9594 1312 | 5036 | 0.012 | 0.5266 | No | ||
| 91 | WNT5A | 22066 5880 | 5078 | 0.011 | 0.5245 | No | ||
| 92 | RSBN1 | 6035 | 5132 | 0.011 | 0.5217 | No | ||
| 93 | RALGPS1 | 2672 6303 | 5269 | 0.010 | 0.5145 | No | ||
| 94 | FBXO33 | 528 | 5314 | 0.009 | 0.5122 | No | ||
| 95 | ARFGEF1 | 9995 | 5411 | 0.009 | 0.5071 | No | ||
| 96 | CUL5 | 7981 | 5664 | 0.007 | 0.4935 | No | ||
| 97 | OSBPL6 | 8277 | 5738 | 0.007 | 0.4896 | No | ||
| 98 | FZD4 | 18193 | 5953 | 0.006 | 0.4781 | No | ||
| 99 | GABRA5 | 17812 | 6151 | 0.005 | 0.4674 | No | ||
| 100 | CPNE8 | 22156 | 6347 | 0.004 | 0.4569 | No | ||
| 101 | DUSP16 | 1004 7699 | 6391 | 0.004 | 0.4546 | No | ||
| 102 | SERTAD2 | 12200 7182 | 6553 | 0.004 | 0.4459 | No | ||
| 103 | PTPN12 | 16597 3502 3586 3665 | 6577 | 0.004 | 0.4447 | No | ||
| 104 | PCDH18 | 15348 | 6683 | 0.003 | 0.4390 | No | ||
| 105 | BASP1 | 7668 | 6701 | 0.003 | 0.4382 | No | ||
| 106 | SPRED2 | 8539 4332 | 6754 | 0.003 | 0.4354 | No | ||
| 107 | DACH1 | 4595 3371 | 6775 | 0.003 | 0.4343 | No | ||
| 108 | PCGF2 | 20270 | 6807 | 0.003 | 0.4327 | No | ||
| 109 | PPP2R5C | 6522 2147 | 6820 | 0.003 | 0.4321 | No | ||
| 110 | CCNT2 | 3993 13079 | 6835 | 0.003 | 0.4314 | No | ||
| 111 | ZBTB11 | 11309 | 6856 | 0.003 | 0.4303 | No | ||
| 112 | SP3 | 5483 | 6954 | 0.003 | 0.4251 | No | ||
| 113 | LRIG1 | 9179 | 6987 | 0.002 | 0.4234 | No | ||
| 114 | RPE | 14232 | 7464 | 0.001 | 0.3975 | No | ||
| 115 | KCNMA1 | 4947 | 7527 | 0.001 | 0.3941 | No | ||
| 116 | GABRA1 | 20492 | 7743 | 0.001 | 0.3825 | No | ||
| 117 | TMEM16D | 19649 | 7771 | 0.001 | 0.3810 | No | ||
| 118 | ATAD2 | 2268 2256 | 7798 | 0.000 | 0.3796 | No | ||
| 119 | GRM5 | 8426 | 8004 | -0.000 | 0.3684 | No | ||
| 120 | UNC50 | 12521 | 8157 | -0.000 | 0.3602 | No | ||
| 121 | PDZRN3 | 17054 | 8511 | -0.001 | 0.3410 | No | ||
| 122 | ADAM10 | 4336 | 8534 | -0.001 | 0.3398 | No | ||
| 123 | RPS6KA5 | 2076 21005 | 8560 | -0.001 | 0.3385 | No | ||
| 124 | VPS13D | 15681 | 8578 | -0.001 | 0.3376 | No | ||
| 125 | AK5 | 6042 | 8951 | -0.002 | 0.3174 | No | ||
| 126 | RIMS3 | 6325 | 8971 | -0.002 | 0.3164 | No | ||
| 127 | ZIC2 | 21927 | 9014 | -0.002 | 0.3141 | No | ||
| 128 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.003 | 0.3087 | No | ||
| 129 | ESRRG | 6447 | 9116 | -0.003 | 0.3088 | No | ||
| 130 | BHLHB5 | 15629 | 9205 | -0.003 | 0.3040 | No | ||
| 131 | SPRYD3 | 522 | 9404 | -0.003 | 0.2933 | No | ||
| 132 | TJP1 | 17803 | 9546 | -0.004 | 0.2857 | No | ||
| 133 | KCNIP4 | 3638 16536 | 9644 | -0.004 | 0.2805 | No | ||
| 134 | NRXN3 | 2153 5196 | 9732 | -0.004 | 0.2758 | No | ||
| 135 | SPIRE2 | 18430 | 9949 | -0.005 | 0.2641 | No | ||
| 136 | SALL1 | 18796 12207 | 9993 | -0.005 | 0.2619 | No | ||
| 137 | LRRTM3 | 19744 | 10004 | -0.005 | 0.2614 | No | ||
| 138 | FIGN | 14575 | 10020 | -0.005 | 0.2606 | No | ||
| 139 | DLL1 | 23119 | 10047 | -0.005 | 0.2593 | No | ||
| 140 | POU4F1 | 21727 | 10137 | -0.005 | 0.2545 | No | ||
| 141 | EPHA7 | 8909 4674 | 10180 | -0.005 | 0.2523 | No | ||
| 142 | SDC2 | 9134 | 10382 | -0.006 | 0.2415 | No | ||
| 143 | DKK2 | 15418 | 10424 | -0.006 | 0.2393 | No | ||
| 144 | CITED2 | 5118 14477 | 10610 | -0.007 | 0.2294 | No | ||
| 145 | ZIC1 | 19040 | 10643 | -0.007 | 0.2277 | No | ||
| 146 | NLGN3 | 10878 | 10956 | -0.008 | 0.2108 | No | ||
| 147 | SEMA4C | 9799 | 11200 | -0.009 | 0.1978 | No | ||
| 148 | GABRG2 | 1259 4748 8996 1331 | 11323 | -0.009 | 0.1912 | No | ||
| 149 | NLGN1 | 15368 | 11329 | -0.009 | 0.1911 | No | ||
| 150 | PDGFC | 7048 | 11413 | -0.010 | 0.1867 | No | ||
| 151 | BHLHB3 | 16935 | 11500 | -0.010 | 0.1822 | No | ||
| 152 | SRPX | 11952 2630 | 11532 | -0.010 | 0.1806 | No | ||
| 153 | NDST1 | 23430 4885 | 11558 | -0.010 | 0.1794 | No | ||
| 154 | PFN2 | 15337 1818 | 11816 | -0.012 | 0.1656 | No | ||
| 155 | UBE2V2 | 12888 | 11851 | -0.012 | 0.1639 | No | ||
| 156 | CDH11 | 18780 3861 8725 | 12014 | -0.013 | 0.1553 | No | ||
| 157 | GPC4 | 24153 | 12024 | -0.013 | 0.1549 | No | ||
| 158 | ARHGAP5 | 4412 8625 | 12055 | -0.013 | 0.1535 | No | ||
| 159 | IRX3 | 18793 | 12071 | -0.013 | 0.1528 | No | ||
| 160 | IL1A | 4915 | 12109 | -0.013 | 0.1510 | No | ||
| 161 | HS3ST3B1 | 7054 | 12311 | -0.015 | 0.1403 | No | ||
| 162 | TTN | 14552 2743 14553 | 12455 | -0.016 | 0.1327 | No | ||
| 163 | FHOD3 | 10340 | 12590 | -0.017 | 0.1256 | No | ||
| 164 | WDFY3 | 7789 3607 16462 | 12763 | -0.018 | 0.1165 | No | ||
| 165 | SNX9 | 23132 | 12859 | -0.019 | 0.1116 | No | ||
| 166 | EFTUD1 | 8316 | 13069 | -0.022 | 0.1006 | No | ||
| 167 | UNC5A | 3288 21631 | 13106 | -0.022 | 0.0989 | No | ||
| 168 | CDH2 | 1963 8727 8726 4508 | 13270 | -0.024 | 0.0903 | No | ||
| 169 | TMEFF1 | 16209 | 13286 | -0.024 | 0.0899 | No | ||
| 170 | PIP5K3 | 14234 | 13388 | -0.026 | 0.0847 | No | ||
| 171 | SNAI1 | 14726 | 13458 | -0.027 | 0.0813 | No | ||
| 172 | MARK1 | 5955 | 13484 | -0.027 | 0.0803 | No | ||
| 173 | HIVEP2 | 9091 | 13748 | -0.032 | 0.0664 | No | ||
| 174 | CAPN3 | 8686 2868 | 13811 | -0.033 | 0.0635 | No | ||
| 175 | DCUN1D1 | 8546 | 13901 | -0.034 | 0.0591 | No | ||
| 176 | ENAH | 4665 8901 | 13909 | -0.035 | 0.0592 | No | ||
| 177 | ANTXR1 | 1079 17082 | 14033 | -0.037 | 0.0530 | No | ||
| 178 | CRSP6 | 19237 | 14206 | -0.042 | 0.0442 | No | ||
| 179 | SRPK2 | 5513 | 14328 | -0.045 | 0.0382 | No | ||
| 180 | CDH13 | 4506 3826 | 14375 | -0.046 | 0.0363 | No | ||
| 181 | OVOL1 | 23983 | 14473 | -0.049 | 0.0317 | No | ||
| 182 | EFNA3 | 15278 | 14497 | -0.050 | 0.0311 | No | ||
| 183 | ZMYND19 | 12484 7402 | 14529 | -0.051 | 0.0301 | No | ||
| 184 | COBLL1 | 2674 6713 | 14649 | -0.057 | 0.0244 | No | ||
| 185 | TBC1D4 | 5579 | 14664 | -0.058 | 0.0244 | No | ||
| 186 | SIN3A | 5442 | 14777 | -0.063 | 0.0191 | No | ||
| 187 | GAD1 | 14993 2690 | 14918 | -0.070 | 0.0124 | No | ||
| 188 | PAIP1 | 21556 | 14971 | -0.073 | 0.0106 | No | ||
| 189 | AR | 24286 | 15108 | -0.083 | 0.0043 | No | ||
| 190 | UBE3A | 1209 1084 1830 | 15158 | -0.086 | 0.0028 | No | ||
| 191 | TRAM2 | 13990 | 15173 | -0.087 | 0.0032 | No | ||
| 192 | SPAG8 | 167 | 15312 | -0.099 | -0.0030 | No | ||
| 193 | RHOT1 | 12227 | 15412 | -0.111 | -0.0069 | No | ||
| 194 | EIF4ENIF1 | 20971 | 15432 | -0.113 | -0.0065 | No | ||
| 195 | KPNA4 | 15321 | 15525 | -0.123 | -0.0099 | No | ||
| 196 | CDC73 | 5657 | 15716 | -0.149 | -0.0182 | No | ||
| 197 | JPH1 | 13994 | 15743 | -0.153 | -0.0176 | No | ||
| 198 | NUDT4 | 19639 | 15934 | -0.187 | -0.0255 | No | ||
| 199 | PUM2 | 2071 8161 | 15940 | -0.188 | -0.0232 | No | ||
| 200 | PPP2R5D | 962 1523 22957 10139 | 16091 | -0.216 | -0.0285 | No | ||
| 201 | CLPX | 19403 | 16183 | -0.233 | -0.0304 | No | ||
| 202 | DHX36 | 15328 | 16290 | -0.259 | -0.0328 | No | ||
| 203 | NMT1 | 20638 | 16432 | -0.292 | -0.0366 | No | ||
| 204 | MTPN | 17183 | 16481 | -0.305 | -0.0351 | No | ||
| 205 | CRSP7 | 18838 | 16534 | -0.319 | -0.0337 | No | ||
| 206 | SFRS3 | 5428 23312 | 16566 | -0.328 | -0.0311 | No | ||
| 207 | TRPS1 | 8195 | 16727 | -0.376 | -0.0348 | No | ||
| 208 | ATP2A2 | 4421 3481 | 16775 | -0.391 | -0.0322 | No | ||
| 209 | XPO7 | 12272 7239 | 16851 | -0.414 | -0.0308 | No | ||
| 210 | E2F6 | 6925 6926 11920 | 16924 | -0.445 | -0.0288 | No | ||
| 211 | DDX3X | 24379 | 17178 | -0.541 | -0.0354 | No | ||
| 212 | OTUB1 | 23795 | 17284 | -0.594 | -0.0333 | No | ||
| 213 | APLP2 | 19190 | 17394 | -0.639 | -0.0308 | No | ||
| 214 | SNRP70 | 1186 | 17417 | -0.653 | -0.0233 | No | ||
| 215 | ATP5A1 | 23505 | 17444 | -0.667 | -0.0159 | No | ||
| 216 | UBE2Q1 | 13662 12851 15537 | 17620 | -0.773 | -0.0152 | No | ||
| 217 | BTG1 | 4458 4457 8663 | 17932 | -0.993 | -0.0190 | No | ||
| 218 | TLE4 | 23720 3697 | 17948 | -1.011 | -0.0064 | No | ||
| 219 | SLC25A26 | 12552 | 17963 | -1.029 | 0.0064 | No | ||
| 220 | ETF1 | 23467 | 18013 | -1.092 | 0.0182 | No | ||
| 221 | CARM1 | 19539 | 18022 | -1.102 | 0.0323 | No |