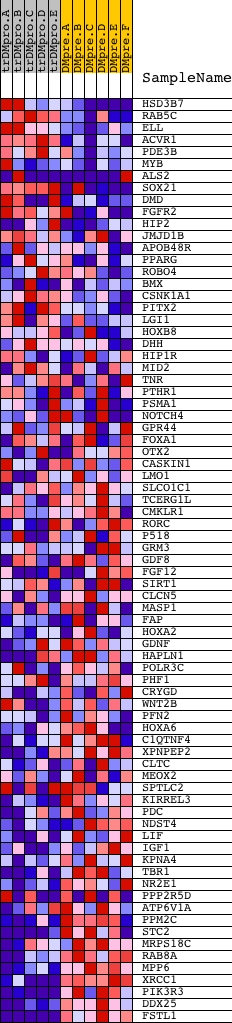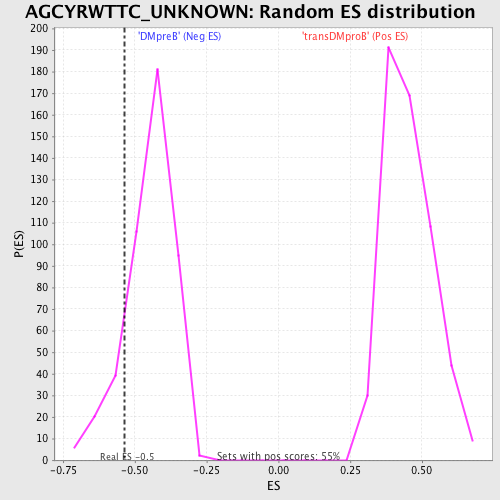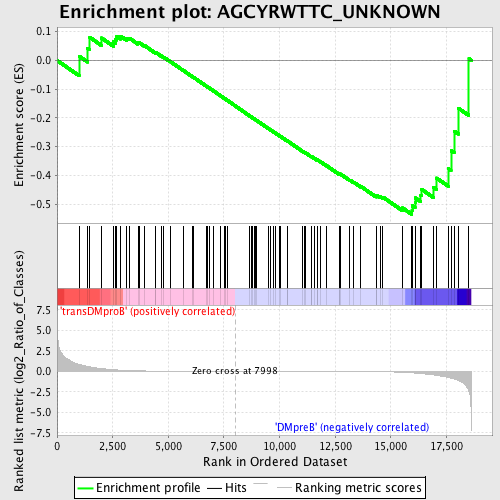
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | AGCYRWTTC_UNKNOWN |
| Enrichment Score (ES) | -0.5356415 |
| Normalized Enrichment Score (NES) | -1.1979219 |
| Nominal p-value | 0.13363029 |
| FDR q-value | 0.7920322 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HSD3B7 | 4158 | 990 | 0.840 | 0.0145 | No | ||
| 2 | RAB5C | 20226 | 1367 | 0.588 | 0.0419 | No | ||
| 3 | ELL | 18586 | 1470 | 0.542 | 0.0802 | No | ||
| 4 | ACVR1 | 4334 | 1996 | 0.324 | 0.0781 | No | ||
| 5 | PDE3B | 5236 | 2535 | 0.194 | 0.0647 | No | ||
| 6 | MYB | 3344 3355 19806 | 2630 | 0.171 | 0.0735 | No | ||
| 7 | ALS2 | 7882 13151 | 2682 | 0.159 | 0.0836 | No | ||
| 8 | SOX21 | 27 10278 2778 | 2857 | 0.127 | 0.0845 | No | ||
| 9 | DMD | 24295 2647 | 3129 | 0.092 | 0.0773 | No | ||
| 10 | FGFR2 | 1917 4722 1119 | 3240 | 0.081 | 0.0779 | No | ||
| 11 | HIP2 | 3626 3485 16838 | 3635 | 0.050 | 0.0607 | No | ||
| 12 | JMJD1B | 23600 | 3693 | 0.047 | 0.0614 | No | ||
| 13 | APOB48R | 18081 | 3936 | 0.036 | 0.0513 | No | ||
| 14 | PPARG | 1151 1144 17319 | 4435 | 0.020 | 0.0261 | No | ||
| 15 | ROBO4 | 7895 3064 | 4440 | 0.020 | 0.0275 | No | ||
| 16 | BMX | 2566 2585 24010 | 4683 | 0.016 | 0.0157 | No | ||
| 17 | CSNK1A1 | 8204 | 4783 | 0.014 | 0.0115 | No | ||
| 18 | PITX2 | 15424 1878 | 5080 | 0.011 | -0.0035 | No | ||
| 19 | LGI1 | 23867 | 5681 | 0.007 | -0.0353 | No | ||
| 20 | HOXB8 | 9111 | 6080 | 0.005 | -0.0564 | No | ||
| 21 | DHH | 4629 | 6117 | 0.005 | -0.0579 | No | ||
| 22 | HIP1R | 16703 | 6705 | 0.003 | -0.0893 | No | ||
| 23 | MID2 | 24232 | 6761 | 0.003 | -0.0920 | No | ||
| 24 | TNR | 5787 | 6852 | 0.003 | -0.0966 | No | ||
| 25 | PTHR1 | 5318 18985 | 7021 | 0.002 | -0.1055 | No | ||
| 26 | PSMA1 | 1627 17669 | 7335 | 0.002 | -0.1222 | No | ||
| 27 | NOTCH4 | 23277 9475 | 7543 | 0.001 | -0.1333 | No | ||
| 28 | GPR44 | 23926 | 7561 | 0.001 | -0.1342 | No | ||
| 29 | FOXA1 | 21060 | 7666 | 0.001 | -0.1397 | No | ||
| 30 | OTX2 | 9519 | 8660 | -0.002 | -0.1931 | No | ||
| 31 | CASKIN1 | 11219 879 | 8744 | -0.002 | -0.1975 | No | ||
| 32 | LMO1 | 17685 | 8803 | -0.002 | -0.2005 | No | ||
| 33 | SLCO1C1 | 17251 | 8859 | -0.002 | -0.2033 | No | ||
| 34 | TCERG1L | 7690 | 8896 | -0.002 | -0.2050 | No | ||
| 35 | CMKLR1 | 16429 | 8975 | -0.002 | -0.2090 | No | ||
| 36 | RORC | 9732 | 9494 | -0.004 | -0.2367 | No | ||
| 37 | P518 | 14624 | 9576 | -0.004 | -0.2408 | No | ||
| 38 | GRM3 | 16929 | 9705 | -0.004 | -0.2473 | No | ||
| 39 | GDF8 | 9411 | 9819 | -0.004 | -0.2531 | No | ||
| 40 | FGF12 | 1723 22621 | 9991 | -0.005 | -0.2619 | No | ||
| 41 | SIRT1 | 19745 | 10054 | -0.005 | -0.2648 | No | ||
| 42 | CLCN5 | 24204 | 10338 | -0.006 | -0.2796 | No | ||
| 43 | MASP1 | 1632 22626 | 10371 | -0.006 | -0.2808 | No | ||
| 44 | FAP | 14577 | 11025 | -0.008 | -0.3154 | No | ||
| 45 | HOXA2 | 17151 | 11113 | -0.009 | -0.3194 | No | ||
| 46 | GDNF | 22514 2275 | 11186 | -0.009 | -0.3225 | No | ||
| 47 | HAPLN1 | 21592 | 11434 | -0.010 | -0.3351 | No | ||
| 48 | POLR3C | 15235 | 11439 | -0.010 | -0.3345 | No | ||
| 49 | PHF1 | 23324 1551 | 11587 | -0.010 | -0.3416 | No | ||
| 50 | CRYGD | 8797 | 11693 | -0.011 | -0.3464 | No | ||
| 51 | WNT2B | 15214 1838 | 11695 | -0.011 | -0.3455 | No | ||
| 52 | PFN2 | 15337 1818 | 11816 | -0.012 | -0.3511 | No | ||
| 53 | HOXA6 | 17150 | 12117 | -0.013 | -0.3662 | No | ||
| 54 | C1QTNF4 | 14953 | 12671 | -0.018 | -0.3946 | No | ||
| 55 | XPNPEP2 | 9309 | 12700 | -0.018 | -0.3946 | No | ||
| 56 | CLTC | 14610 | 12730 | -0.018 | -0.3947 | No | ||
| 57 | MEOX2 | 21285 | 13158 | -0.023 | -0.4159 | No | ||
| 58 | SPTLC2 | 5505 5506 | 13339 | -0.025 | -0.4236 | No | ||
| 59 | KIRREL3 | 12571 | 13651 | -0.030 | -0.4380 | No | ||
| 60 | PDC | 4046 14102 | 14337 | -0.045 | -0.4713 | No | ||
| 61 | NDST4 | 12259 7228 | 14367 | -0.046 | -0.4692 | No | ||
| 62 | LIF | 956 667 20960 | 14523 | -0.051 | -0.4734 | No | ||
| 63 | IGF1 | 3352 9156 3409 | 14645 | -0.057 | -0.4753 | No | ||
| 64 | KPNA4 | 15321 | 15525 | -0.123 | -0.5128 | No | ||
| 65 | TBR1 | 83 | 15950 | -0.190 | -0.5203 | Yes | ||
| 66 | NR2E1 | 19780 | 15961 | -0.192 | -0.5054 | Yes | ||
| 67 | PPP2R5D | 962 1523 22957 10139 | 16091 | -0.216 | -0.4948 | Yes | ||
| 68 | ATP6V1A | 8638 | 16105 | -0.218 | -0.4780 | Yes | ||
| 69 | PPM2C | 491 | 16348 | -0.271 | -0.4691 | Yes | ||
| 70 | STC2 | 5526 | 16391 | -0.282 | -0.4485 | Yes | ||
| 71 | MRPS18C | 12728 | 16912 | -0.441 | -0.4409 | Yes | ||
| 72 | RAB8A | 9380 5091 | 17052 | -0.490 | -0.4088 | Yes | ||
| 73 | MPP6 | 17448 | 17598 | -0.760 | -0.3766 | Yes | ||
| 74 | XRCC1 | 173 | 17707 | -0.843 | -0.3143 | Yes | ||
| 75 | PIK3R3 | 5248 | 17847 | -0.935 | -0.2461 | Yes | ||
| 76 | DDX25 | 19184 | 18033 | -1.115 | -0.1659 | Yes | ||
| 77 | FSTL1 | 22765 | 18512 | -2.438 | 0.0056 | Yes |