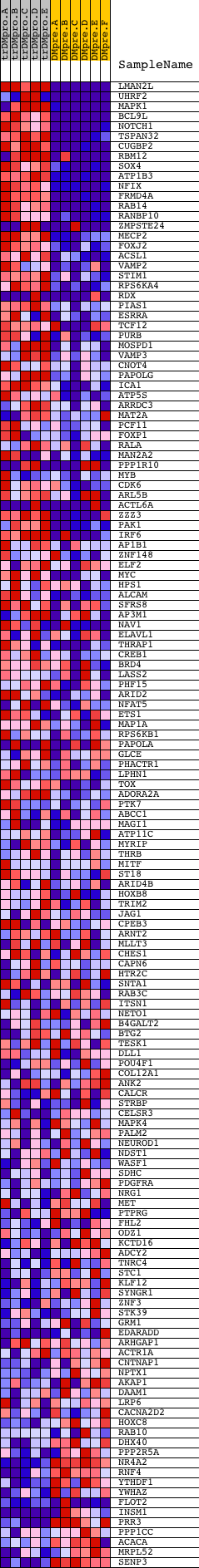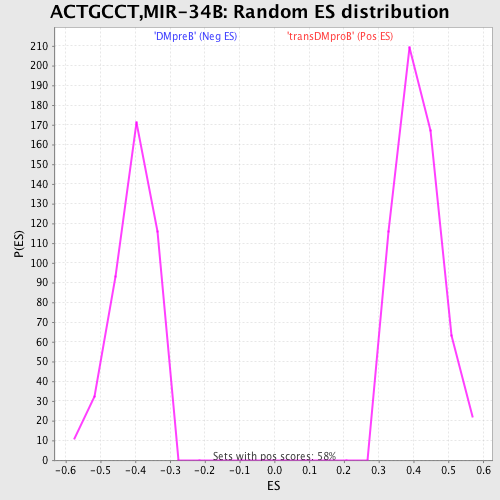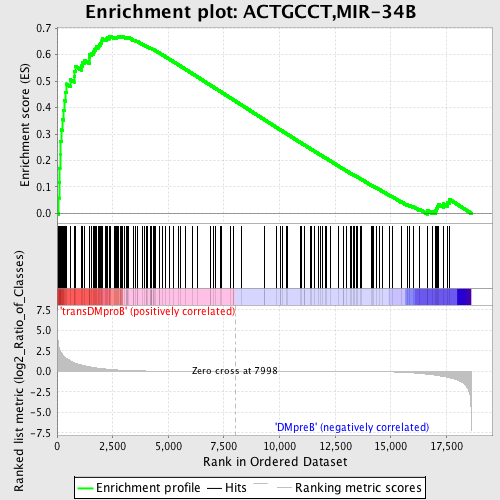
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | ACTGCCT,MIR-34B |
| Enrichment Score (ES) | 0.67015404 |
| Normalized Enrichment Score (NES) | 1.6148925 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.029138329 |
| FWER p-Value | 0.177 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LMAN2L | 5670 10070 5669 | 79 | 3.002 | 0.0582 | Yes | ||
| 2 | UHRF2 | 3731 4262 8452 23887 | 88 | 2.827 | 0.1165 | Yes | ||
| 3 | MAPK1 | 1642 11167 | 108 | 2.667 | 0.1709 | Yes | ||
| 4 | BCL9L | 19475 | 131 | 2.500 | 0.2217 | Yes | ||
| 5 | NOTCH1 | 14649 | 135 | 2.489 | 0.2733 | Yes | ||
| 6 | TSPAN32 | 18002 | 192 | 2.210 | 0.3162 | Yes | ||
| 7 | CUGBP2 | 4687 8923 | 251 | 2.014 | 0.3549 | Yes | ||
| 8 | RBM12 | 7980 2710 13280 | 299 | 1.837 | 0.3906 | Yes | ||
| 9 | SOX4 | 5479 | 318 | 1.801 | 0.4271 | Yes | ||
| 10 | ATP1B3 | 19037 | 385 | 1.629 | 0.4574 | Yes | ||
| 11 | NFIX | 9458 | 409 | 1.590 | 0.4892 | Yes | ||
| 12 | FRMD4A | 5557 | 603 | 1.272 | 0.5052 | Yes | ||
| 13 | RAB14 | 12685 | 764 | 1.035 | 0.5180 | Yes | ||
| 14 | RANBP10 | 18764 | 790 | 1.006 | 0.5376 | Yes | ||
| 15 | ZMPSTE24 | 6061 6060 10509 15769 | 828 | 0.968 | 0.5557 | Yes | ||
| 16 | MECP2 | 5088 | 1090 | 0.766 | 0.5575 | Yes | ||
| 17 | FOXJ2 | 17294 | 1131 | 0.732 | 0.5706 | Yes | ||
| 18 | ACSL1 | 8948 | 1226 | 0.663 | 0.5793 | Yes | ||
| 19 | VAMP2 | 20826 | 1445 | 0.551 | 0.5789 | Yes | ||
| 20 | STIM1 | 18164 | 1447 | 0.550 | 0.5903 | Yes | ||
| 21 | RPS6KA4 | 23804 | 1464 | 0.544 | 0.6008 | Yes | ||
| 22 | RDX | 9712 5371 | 1562 | 0.495 | 0.6058 | Yes | ||
| 23 | PIAS1 | 7126 | 1633 | 0.464 | 0.6116 | Yes | ||
| 24 | ESRRA | 23802 | 1665 | 0.453 | 0.6194 | Yes | ||
| 25 | TCF12 | 10044 3125 | 1741 | 0.416 | 0.6240 | Yes | ||
| 26 | PURB | 5335 | 1757 | 0.410 | 0.6317 | Yes | ||
| 27 | MOSPD1 | 12868 12869 7672 | 1880 | 0.363 | 0.6326 | Yes | ||
| 28 | VAMP3 | 15660 | 1886 | 0.361 | 0.6399 | Yes | ||
| 29 | CNOT4 | 7010 1022 | 1958 | 0.336 | 0.6430 | Yes | ||
| 30 | PAPOLG | 20512 | 1978 | 0.332 | 0.6489 | Yes | ||
| 31 | ICA1 | 17217 | 2013 | 0.319 | 0.6537 | Yes | ||
| 32 | ATP5S | 21251 | 2024 | 0.316 | 0.6597 | Yes | ||
| 33 | ARRDC3 | 4191 | 2168 | 0.274 | 0.6577 | Yes | ||
| 34 | MAT2A | 10553 10554 6099 | 2233 | 0.259 | 0.6596 | Yes | ||
| 35 | PCF11 | 121 | 2247 | 0.256 | 0.6642 | Yes | ||
| 36 | FOXP1 | 4242 | 2341 | 0.235 | 0.6641 | Yes | ||
| 37 | RALA | 21541 | 2350 | 0.234 | 0.6685 | Yes | ||
| 38 | MAN2A2 | 4696 8931 | 2405 | 0.221 | 0.6702 | Yes | ||
| 39 | PPP1R10 | 6972 11960 6971 | 2593 | 0.179 | 0.6637 | No | ||
| 40 | MYB | 3344 3355 19806 | 2630 | 0.171 | 0.6654 | No | ||
| 41 | CDK6 | 16600 | 2683 | 0.159 | 0.6658 | No | ||
| 42 | ARL5B | 15115 | 2727 | 0.149 | 0.6666 | No | ||
| 43 | ACTL6A | 12136 13068 16662 3457 3554 | 2767 | 0.142 | 0.6675 | No | ||
| 44 | ZZZ3 | 15389 8445 4253 8444 1761 | 2842 | 0.130 | 0.6662 | No | ||
| 45 | PAK1 | 9527 | 2858 | 0.127 | 0.6680 | No | ||
| 46 | IRF6 | 4013 12011 4048 7024 | 2914 | 0.116 | 0.6674 | No | ||
| 47 | AP1B1 | 8599 | 2917 | 0.116 | 0.6697 | No | ||
| 48 | ZNF148 | 5949 | 3036 | 0.101 | 0.6655 | No | ||
| 49 | ELF2 | 1795 1856 12785 1882 | 3105 | 0.095 | 0.6637 | No | ||
| 50 | MYC | 22465 9435 | 3120 | 0.093 | 0.6649 | No | ||
| 51 | HPS1 | 23671 | 3158 | 0.089 | 0.6648 | No | ||
| 52 | ALCAM | 4367 | 3162 | 0.088 | 0.6664 | No | ||
| 53 | SFRS8 | 10543 6089 | 3220 | 0.082 | 0.6651 | No | ||
| 54 | AP3M1 | 7059 | 3440 | 0.063 | 0.6545 | No | ||
| 55 | NAV1 | 3978 5681 | 3448 | 0.063 | 0.6555 | No | ||
| 56 | ELAVL1 | 4888 | 3527 | 0.057 | 0.6524 | No | ||
| 57 | THRAP1 | 20309 | 3592 | 0.053 | 0.6501 | No | ||
| 58 | CREB1 | 3990 8782 4558 4093 | 3625 | 0.050 | 0.6494 | No | ||
| 59 | BRD4 | 7164 1609 1628 1598 1552 1601 12176 1630 1501 | 3852 | 0.039 | 0.6380 | No | ||
| 60 | LASS2 | 15507 13366 | 3943 | 0.035 | 0.6338 | No | ||
| 61 | PHF15 | 20467 8050 | 4014 | 0.032 | 0.6307 | No | ||
| 62 | ARID2 | 8062 13378 | 4025 | 0.032 | 0.6308 | No | ||
| 63 | NFAT5 | 3921 7037 12036 | 4081 | 0.030 | 0.6285 | No | ||
| 64 | ETS1 | 10715 6230 3135 | 4196 | 0.026 | 0.6228 | No | ||
| 65 | MAP1A | 5124 | 4197 | 0.026 | 0.6234 | No | ||
| 66 | RPS6KB1 | 7815 1207 13040 | 4229 | 0.026 | 0.6222 | No | ||
| 67 | PAPOLA | 5261 9585 | 4236 | 0.025 | 0.6225 | No | ||
| 68 | GLCE | 13526 | 4246 | 0.025 | 0.6225 | No | ||
| 69 | PHACTR1 | 21657 3247 | 4325 | 0.023 | 0.6188 | No | ||
| 70 | LPHN1 | 6850 174 | 4337 | 0.023 | 0.6186 | No | ||
| 71 | TOX | 15943 | 4359 | 0.023 | 0.6180 | No | ||
| 72 | ADORA2A | 8557 | 4438 | 0.020 | 0.6142 | No | ||
| 73 | PTK7 | 22959 | 4584 | 0.017 | 0.6067 | No | ||
| 74 | ABCC1 | 1668 5087 | 4586 | 0.017 | 0.6070 | No | ||
| 75 | MAGI1 | 19828 4816 1179 | 4716 | 0.015 | 0.6003 | No | ||
| 76 | ATP11C | 6813 24146 | 4888 | 0.013 | 0.5913 | No | ||
| 77 | MYRIP | 19269 | 5062 | 0.011 | 0.5822 | No | ||
| 78 | THRB | 10173 | 5211 | 0.010 | 0.5744 | No | ||
| 79 | MITF | 17349 | 5449 | 0.008 | 0.5617 | No | ||
| 80 | ST18 | 14298 | 5526 | 0.008 | 0.5578 | No | ||
| 81 | ARID4B | 21705 3233 | 5765 | 0.007 | 0.5450 | No | ||
| 82 | HOXB8 | 9111 | 6080 | 0.005 | 0.5281 | No | ||
| 83 | TRIM2 | 13485 8157 | 6310 | 0.005 | 0.5158 | No | ||
| 84 | JAG1 | 14415 | 6890 | 0.003 | 0.4845 | No | ||
| 85 | CPEB3 | 5539 | 6900 | 0.003 | 0.4841 | No | ||
| 86 | ARNT2 | 17769 | 7049 | 0.002 | 0.4761 | No | ||
| 87 | MLLT3 | 2359 15847 2413 | 7133 | 0.002 | 0.4717 | No | ||
| 88 | CHES1 | 21007 11200 | 7323 | 0.002 | 0.4615 | No | ||
| 89 | CAPN6 | 24041 | 7380 | 0.001 | 0.4585 | No | ||
| 90 | HTR2C | 24230 | 7789 | 0.000 | 0.4364 | No | ||
| 91 | SNTA1 | 14386 | 7919 | 0.000 | 0.4294 | No | ||
| 92 | RAB3C | 21351 | 7947 | 0.000 | 0.4280 | No | ||
| 93 | ITSN1 | 9196 | 8307 | -0.001 | 0.4085 | No | ||
| 94 | NETO1 | 6372 1970 | 9321 | -0.003 | 0.3538 | No | ||
| 95 | B4GALT2 | 11990 | 9331 | -0.003 | 0.3533 | No | ||
| 96 | BTG2 | 13832 | 9851 | -0.005 | 0.3253 | No | ||
| 97 | TESK1 | 16233 | 10046 | -0.005 | 0.3149 | No | ||
| 98 | DLL1 | 23119 | 10047 | -0.005 | 0.3150 | No | ||
| 99 | POU4F1 | 21727 | 10137 | -0.005 | 0.3103 | No | ||
| 100 | COL12A1 | 3143 19054 | 10325 | -0.006 | 0.3003 | No | ||
| 101 | ANK2 | 4275 | 10350 | -0.006 | 0.2992 | No | ||
| 102 | CALCR | 17229 | 10932 | -0.008 | 0.2679 | No | ||
| 103 | STRBP | 14601 5496 | 10962 | -0.008 | 0.2665 | No | ||
| 104 | CELSR3 | 19305 8419 | 11117 | -0.009 | 0.2583 | No | ||
| 105 | MAPK4 | 23409 | 11140 | -0.009 | 0.2573 | No | ||
| 106 | PALM2 | 10817 2368 | 11406 | -0.010 | 0.2431 | No | ||
| 107 | NEUROD1 | 14550 | 11436 | -0.010 | 0.2418 | No | ||
| 108 | NDST1 | 23430 4885 | 11558 | -0.010 | 0.2354 | No | ||
| 109 | WASF1 | 13514 | 11763 | -0.011 | 0.2246 | No | ||
| 110 | SDHC | 7248 12279 | 11846 | -0.012 | 0.2204 | No | ||
| 111 | PDGFRA | 16824 | 11924 | -0.012 | 0.2165 | No | ||
| 112 | NRG1 | 3885 | 12063 | -0.013 | 0.2093 | No | ||
| 113 | MET | 17520 | 12124 | -0.013 | 0.2063 | No | ||
| 114 | PTPRG | 22097 3151 | 12299 | -0.015 | 0.1972 | No | ||
| 115 | FHL2 | 13966 | 12656 | -0.017 | 0.1783 | No | ||
| 116 | ODZ1 | 24168 | 12857 | -0.019 | 0.1679 | No | ||
| 117 | KCTD16 | 11803 | 12873 | -0.020 | 0.1675 | No | ||
| 118 | ADCY2 | 21412 | 13026 | -0.021 | 0.1597 | No | ||
| 119 | TNRC4 | 15514 | 13167 | -0.023 | 0.1526 | No | ||
| 120 | STC1 | 21974 | 13241 | -0.024 | 0.1491 | No | ||
| 121 | KLF12 | 4960 2637 21732 9228 | 13305 | -0.025 | 0.1462 | No | ||
| 122 | SYNGR1 | 22420 2290 | 13358 | -0.025 | 0.1439 | No | ||
| 123 | ZNF3 | 7094 3516 12097 | 13451 | -0.027 | 0.1395 | No | ||
| 124 | STK39 | 14570 | 13512 | -0.028 | 0.1368 | No | ||
| 125 | GRM1 | 19822 | 13632 | -0.030 | 0.1310 | No | ||
| 126 | EDARADD | 5056 3282 3278 | 13681 | -0.030 | 0.1290 | No | ||
| 127 | ARHGAP1 | 6001 10448 | 14130 | -0.040 | 0.1056 | No | ||
| 128 | ACTR1A | 23653 | 14181 | -0.041 | 0.1037 | No | ||
| 129 | CNTNAP1 | 11987 | 14234 | -0.042 | 0.1018 | No | ||
| 130 | NPTX1 | 20126 | 14338 | -0.045 | 0.0972 | No | ||
| 131 | AKAP1 | 4364 1326 1286 20299 | 14487 | -0.050 | 0.0902 | No | ||
| 132 | DAAM1 | 5536 | 14638 | -0.057 | 0.0832 | No | ||
| 133 | LRP6 | 9286 | 14924 | -0.070 | 0.0693 | No | ||
| 134 | CACNA2D2 | 7154 | 15056 | -0.079 | 0.0638 | No | ||
| 135 | HOXC8 | 4865 | 15463 | -0.116 | 0.0442 | No | ||
| 136 | RAB10 | 2090 21119 | 15734 | -0.152 | 0.0328 | No | ||
| 137 | DHX40 | 20307 | 15843 | -0.171 | 0.0305 | No | ||
| 138 | PPP2R5A | 10386 | 16029 | -0.204 | 0.0247 | No | ||
| 139 | NR4A2 | 5203 | 16292 | -0.259 | 0.0159 | No | ||
| 140 | RNF4 | 5385 9729 | 16637 | -0.351 | 0.0046 | No | ||
| 141 | YTHDF1 | 14311 | 16646 | -0.355 | 0.0115 | No | ||
| 142 | YWHAZ | 10370 | 16871 | -0.421 | 0.0081 | No | ||
| 143 | FLOT2 | 1247 8975 1430 | 17014 | -0.476 | 0.0104 | No | ||
| 144 | INSM1 | 14815 | 17045 | -0.486 | 0.0189 | No | ||
| 145 | PRR3 | 22995 | 17080 | -0.500 | 0.0274 | No | ||
| 146 | PPP1CC | 9609 5283 | 17139 | -0.524 | 0.0352 | No | ||
| 147 | ACACA | 309 4209 | 17370 | -0.627 | 0.0358 | No | ||
| 148 | MRPL52 | 22019 | 17561 | -0.741 | 0.0409 | No | ||
| 149 | SENP3 | 20387 | 17629 | -0.779 | 0.0534 | No |