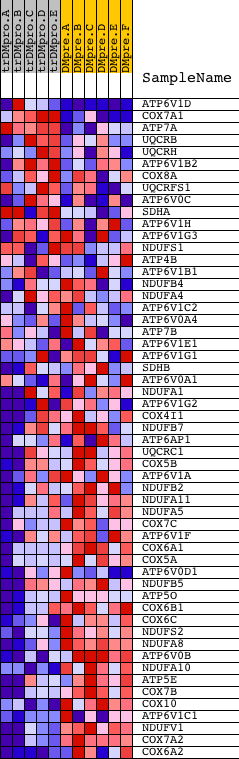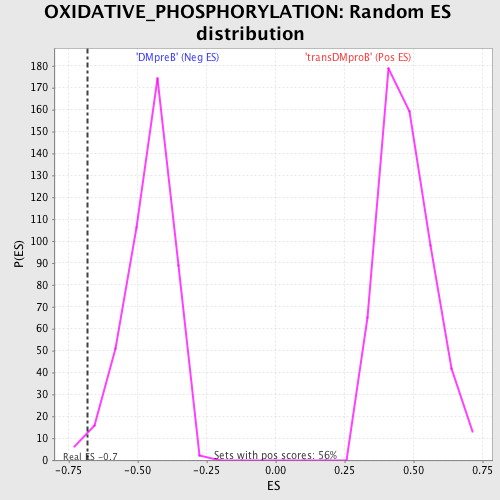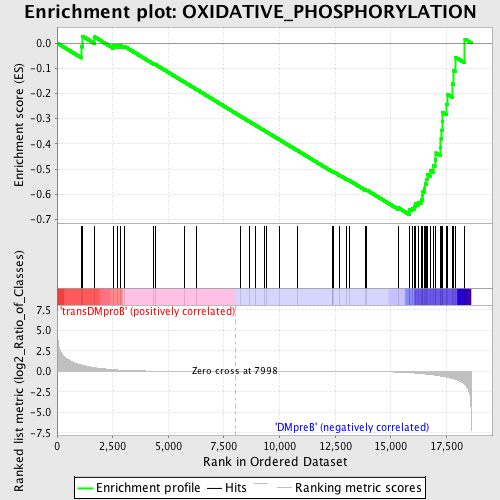
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.6803206 |
| Normalized Enrichment Score (NES) | -1.4819703 |
| Nominal p-value | 0.015765766 |
| FDR q-value | 0.2716074 |
| FWER p-Value | 0.962 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP6V1D | 7872 | 1105 | 0.757 | -0.0144 | No | ||
| 2 | COX7A1 | 4554 | 1138 | 0.728 | 0.0273 | No | ||
| 3 | ATP7A | 4425 | 1685 | 0.441 | 0.0242 | No | ||
| 4 | UQCRB | 12545 | 2529 | 0.195 | -0.0096 | No | ||
| 5 | UQCRH | 12378 | 2701 | 0.155 | -0.0096 | No | ||
| 6 | ATP6V1B2 | 18599 | 2837 | 0.132 | -0.0090 | No | ||
| 7 | COX8A | 8777 | 3014 | 0.104 | -0.0123 | No | ||
| 8 | UQCRFS1 | 21499 | 4342 | 0.023 | -0.0824 | No | ||
| 9 | ATP6V0C | 8643 | 4424 | 0.021 | -0.0855 | No | ||
| 10 | SDHA | 12436 | 5746 | 0.007 | -0.1563 | No | ||
| 11 | ATP6V1H | 14301 | 6273 | 0.005 | -0.1843 | No | ||
| 12 | ATP6V1G3 | 14107 | 8243 | -0.001 | -0.2903 | No | ||
| 13 | NDUFS1 | 13940 | 8627 | -0.001 | -0.3109 | No | ||
| 14 | ATP4B | 18928 | 8912 | -0.002 | -0.3261 | No | ||
| 15 | ATP6V1B1 | 8499 | 9323 | -0.003 | -0.3480 | No | ||
| 16 | NDUFB4 | 22596 7541 | 9396 | -0.003 | -0.3516 | No | ||
| 17 | NDUFA4 | 9451 | 9982 | -0.005 | -0.3829 | No | ||
| 18 | ATP6V1C2 | 21098 | 10797 | -0.007 | -0.4263 | No | ||
| 19 | ATP6V0A4 | 8934 | 12382 | -0.015 | -0.5107 | No | ||
| 20 | ATP7B | 18918 4426 4427 8641 | 12385 | -0.015 | -0.5099 | No | ||
| 21 | ATP6V1E1 | 4423 | 12441 | -0.016 | -0.5119 | No | ||
| 22 | ATP6V1G1 | 12322 | 12683 | -0.018 | -0.5238 | No | ||
| 23 | SDHB | 2348 12566 14880 | 13027 | -0.021 | -0.5410 | No | ||
| 24 | ATP6V0A1 | 8640 4424 1197 | 13131 | -0.022 | -0.5452 | No | ||
| 25 | NDUFA1 | 24170 | 13863 | -0.034 | -0.5826 | No | ||
| 26 | ATP6V1G2 | 23257 1534 | 13924 | -0.035 | -0.5838 | No | ||
| 27 | COX4I1 | 18444 | 15341 | -0.102 | -0.6540 | No | ||
| 28 | NDUFB7 | 12429 | 15831 | -0.168 | -0.6703 | Yes | ||
| 29 | ATP6AP1 | 24296 | 15856 | -0.173 | -0.6613 | Yes | ||
| 30 | UQCRC1 | 10259 | 15952 | -0.190 | -0.6551 | Yes | ||
| 31 | COX5B | 4552 | 16048 | -0.208 | -0.6478 | Yes | ||
| 32 | ATP6V1A | 8638 | 16105 | -0.218 | -0.6378 | Yes | ||
| 33 | NDUFB2 | 7542 | 16252 | -0.250 | -0.6308 | Yes | ||
| 34 | NDUFA11 | 12832 | 16359 | -0.273 | -0.6202 | Yes | ||
| 35 | NDUFA5 | 1076 7544 | 16414 | -0.286 | -0.6060 | Yes | ||
| 36 | COX7C | 8776 | 16420 | -0.288 | -0.5891 | Yes | ||
| 37 | ATP6V1F | 12291 | 16525 | -0.316 | -0.5759 | Yes | ||
| 38 | COX6A1 | 8774 4553 | 16536 | -0.320 | -0.5573 | Yes | ||
| 39 | COX5A | 19431 | 16614 | -0.343 | -0.5410 | Yes | ||
| 40 | ATP6V0D1 | 3895 8639 3920 | 16635 | -0.351 | -0.5212 | Yes | ||
| 41 | NDUFB5 | 12277 | 16767 | -0.389 | -0.5050 | Yes | ||
| 42 | ATP5O | 22539 | 16907 | -0.437 | -0.4864 | Yes | ||
| 43 | COX6B1 | 4283 8479 | 16988 | -0.469 | -0.4627 | Yes | ||
| 44 | COX6C | 8775 | 17029 | -0.479 | -0.4363 | Yes | ||
| 45 | NDUFS2 | 4089 13760 | 17217 | -0.557 | -0.4131 | Yes | ||
| 46 | NDUFA8 | 14605 | 17254 | -0.578 | -0.3805 | Yes | ||
| 47 | ATP6V0B | 15786 | 17293 | -0.597 | -0.3470 | Yes | ||
| 48 | NDUFA10 | 7420 | 17313 | -0.605 | -0.3119 | Yes | ||
| 49 | ATP5E | 14321 | 17331 | -0.611 | -0.2764 | Yes | ||
| 50 | COX7B | 7260 | 17497 | -0.695 | -0.2438 | Yes | ||
| 51 | COX10 | 20408 | 17534 | -0.719 | -0.2028 | Yes | ||
| 52 | ATP6V1C1 | 22487 12329 | 17771 | -0.895 | -0.1622 | Yes | ||
| 53 | NDUFV1 | 23953 | 17810 | -0.917 | -0.1095 | Yes | ||
| 54 | COX7A2 | 19053 | 17907 | -0.978 | -0.0563 | Yes | ||
| 55 | COX6A2 | 17605 | 18332 | -1.583 | 0.0153 | Yes |