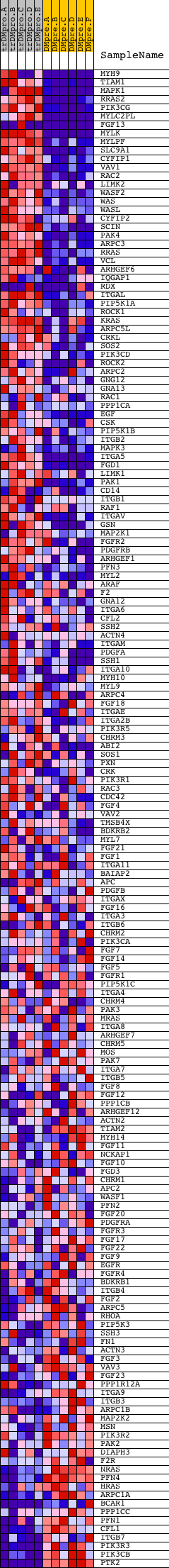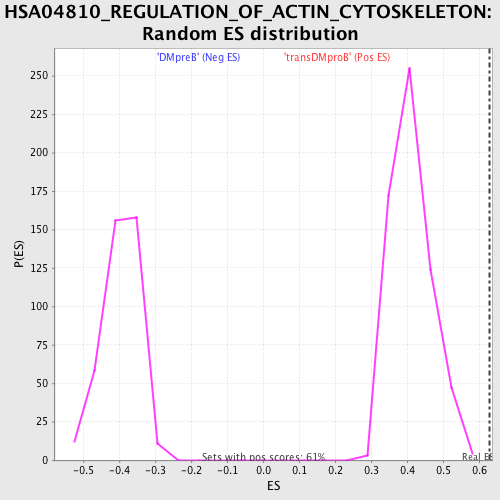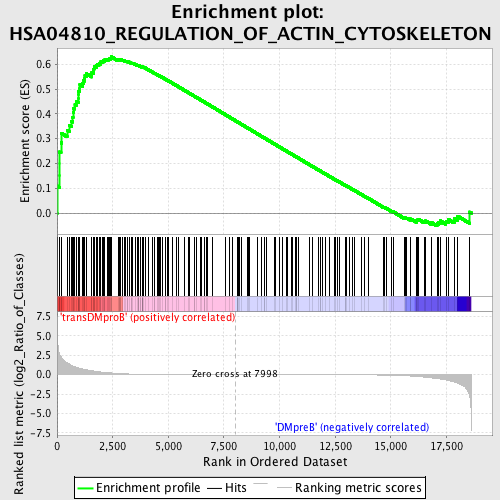
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | HSA04810_REGULATION_OF_ACTIN_CYTOSKELETON |
| Enrichment Score (ES) | 0.62954473 |
| Normalized Enrichment Score (NES) | 1.5307636 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14475422 |
| FWER p-Value | 0.75 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MYH9 | 2252 2244 | 4 | 6.155 | 0.1093 | Yes | ||
| 2 | TIAM1 | 22547 | 106 | 2.676 | 0.1515 | Yes | ||
| 3 | MAPK1 | 1642 11167 | 108 | 2.667 | 0.1989 | Yes | ||
| 4 | RRAS2 | 12431 | 109 | 2.666 | 0.2463 | Yes | ||
| 5 | PIK3CG | 6635 | 176 | 2.268 | 0.2831 | Yes | ||
| 6 | MYLC2PL | 16670 | 196 | 2.203 | 0.3213 | Yes | ||
| 7 | FGF13 | 4720 8963 2627 | 450 | 1.532 | 0.3348 | Yes | ||
| 8 | MYLK | 22778 4213 | 547 | 1.366 | 0.3539 | Yes | ||
| 9 | MYLPF | 18069 | 650 | 1.191 | 0.3696 | Yes | ||
| 10 | SLC9A1 | 16053 | 703 | 1.103 | 0.3864 | Yes | ||
| 11 | CYFIP1 | 9814 | 718 | 1.089 | 0.4051 | Yes | ||
| 12 | VAV1 | 23173 | 746 | 1.053 | 0.4223 | Yes | ||
| 13 | RAC2 | 22217 | 803 | 0.996 | 0.4370 | Yes | ||
| 14 | LIMK2 | 9278 1296 | 857 | 0.943 | 0.4509 | Yes | ||
| 15 | WASF2 | 6326 | 953 | 0.865 | 0.4612 | Yes | ||
| 16 | WAS | 24193 | 958 | 0.862 | 0.4763 | Yes | ||
| 17 | WASL | 17203 | 963 | 0.859 | 0.4914 | Yes | ||
| 18 | CYFIP2 | 20485 8045 | 1018 | 0.819 | 0.5030 | Yes | ||
| 19 | SCIN | 21079 | 1024 | 0.816 | 0.5173 | Yes | ||
| 20 | PAK4 | 17909 | 1147 | 0.717 | 0.5234 | Yes | ||
| 21 | ARPC3 | 12109 | 1198 | 0.682 | 0.5329 | Yes | ||
| 22 | RRAS | 18256 | 1216 | 0.672 | 0.5439 | Yes | ||
| 23 | VCL | 22083 | 1251 | 0.653 | 0.5537 | Yes | ||
| 24 | ARHGEF6 | 13297 13104 | 1312 | 0.617 | 0.5614 | Yes | ||
| 25 | IQGAP1 | 6619 | 1553 | 0.499 | 0.5573 | Yes | ||
| 26 | RDX | 9712 5371 | 1562 | 0.495 | 0.5656 | Yes | ||
| 27 | ITGAL | 9187 | 1625 | 0.469 | 0.5706 | Yes | ||
| 28 | PIP5K1A | 23708 | 1642 | 0.461 | 0.5780 | Yes | ||
| 29 | ROCK1 | 5386 | 1659 | 0.454 | 0.5852 | Yes | ||
| 30 | KRAS | 9247 | 1683 | 0.444 | 0.5918 | Yes | ||
| 31 | ARPC5L | 15017 | 1775 | 0.401 | 0.5940 | Yes | ||
| 32 | CRKL | 4560 | 1800 | 0.391 | 0.5997 | Yes | ||
| 33 | SOS2 | 21049 | 1883 | 0.362 | 0.6017 | Yes | ||
| 34 | PIK3CD | 9563 | 1939 | 0.342 | 0.6048 | Yes | ||
| 35 | ROCK2 | 21309 5387 | 1969 | 0.334 | 0.6092 | Yes | ||
| 36 | ARPC2 | 14225 | 2033 | 0.314 | 0.6114 | Yes | ||
| 37 | GNG12 | 4789 | 2082 | 0.298 | 0.6141 | Yes | ||
| 38 | GNA13 | 20617 | 2135 | 0.285 | 0.6163 | Yes | ||
| 39 | RAC1 | 16302 | 2149 | 0.281 | 0.6206 | Yes | ||
| 40 | PPP1CA | 9608 3756 914 909 | 2257 | 0.252 | 0.6193 | Yes | ||
| 41 | EGF | 15169 | 2310 | 0.241 | 0.6207 | Yes | ||
| 42 | CSK | 8805 | 2369 | 0.230 | 0.6217 | Yes | ||
| 43 | PIP5K1B | 5251 1841 | 2416 | 0.219 | 0.6231 | Yes | ||
| 44 | ITGB2 | 19978 | 2429 | 0.216 | 0.6263 | Yes | ||
| 45 | MAPK3 | 6458 11170 | 2440 | 0.214 | 0.6295 | Yes | ||
| 46 | ITGA5 | 22105 | 2740 | 0.146 | 0.6159 | No | ||
| 47 | FGD1 | 24228 | 2792 | 0.137 | 0.6156 | No | ||
| 48 | LIMK1 | 16350 | 2800 | 0.137 | 0.6177 | No | ||
| 49 | PAK1 | 9527 | 2858 | 0.127 | 0.6168 | No | ||
| 50 | CD14 | 23452 | 2861 | 0.126 | 0.6190 | No | ||
| 51 | ITGB1 | 3872 18411 | 2943 | 0.113 | 0.6166 | No | ||
| 52 | RAF1 | 17035 | 3038 | 0.101 | 0.6133 | No | ||
| 53 | ITGAV | 4931 | 3093 | 0.096 | 0.6121 | No | ||
| 54 | GSN | 2784 | 3156 | 0.090 | 0.6103 | No | ||
| 55 | MAP2K1 | 19082 | 3174 | 0.087 | 0.6109 | No | ||
| 56 | FGFR2 | 1917 4722 1119 | 3240 | 0.081 | 0.6088 | No | ||
| 57 | PDGFRB | 23539 | 3339 | 0.072 | 0.6048 | No | ||
| 58 | ARHGEF1 | 1952 18343 3996 | 3364 | 0.069 | 0.6047 | No | ||
| 59 | PFN3 | 7968 | 3402 | 0.066 | 0.6039 | No | ||
| 60 | MYL2 | 16713 | 3500 | 0.059 | 0.5997 | No | ||
| 61 | ARAF | 24367 | 3602 | 0.052 | 0.5952 | No | ||
| 62 | F2 | 14524 | 3648 | 0.049 | 0.5936 | No | ||
| 63 | GNA12 | 9023 | 3653 | 0.049 | 0.5943 | No | ||
| 64 | ITGA6 | 4930 | 3674 | 0.048 | 0.5940 | No | ||
| 65 | CFL2 | 2155 21069 | 3732 | 0.045 | 0.5917 | No | ||
| 66 | SSH2 | 6202 | 3751 | 0.044 | 0.5915 | No | ||
| 67 | ACTN4 | 17905 1798 1983 | 3815 | 0.041 | 0.5889 | No | ||
| 68 | ITGAM | 9188 | 3824 | 0.041 | 0.5891 | No | ||
| 69 | PDGFA | 5237 | 3830 | 0.040 | 0.5896 | No | ||
| 70 | SSH1 | 10540 | 3858 | 0.039 | 0.5888 | No | ||
| 71 | ITGA10 | 15493 | 3899 | 0.038 | 0.5873 | No | ||
| 72 | MYH10 | 8077 | 3952 | 0.035 | 0.5851 | No | ||
| 73 | MYL9 | 14767 2659 | 4114 | 0.029 | 0.5769 | No | ||
| 74 | ARPC4 | 12642 | 4293 | 0.024 | 0.5677 | No | ||
| 75 | FGF18 | 4721 1351 | 4370 | 0.022 | 0.5640 | No | ||
| 76 | ITGAE | 20791 1481 1397 1380 1278 1238 1359 | 4393 | 0.021 | 0.5631 | No | ||
| 77 | ITGA2B | 9186 | 4503 | 0.019 | 0.5576 | No | ||
| 78 | PIK3R5 | 11507 | 4539 | 0.018 | 0.5560 | No | ||
| 79 | CHRM3 | 21547 | 4622 | 0.017 | 0.5519 | No | ||
| 80 | ABI2 | 4037 6830 | 4664 | 0.016 | 0.5499 | No | ||
| 81 | SOS1 | 5476 | 4722 | 0.015 | 0.5471 | No | ||
| 82 | PXN | 5339 3573 | 4852 | 0.013 | 0.5403 | No | ||
| 83 | CRK | 4559 1249 | 4892 | 0.013 | 0.5384 | No | ||
| 84 | PIK3R1 | 3170 | 4940 | 0.013 | 0.5361 | No | ||
| 85 | RAC3 | 20561 | 4987 | 0.012 | 0.5338 | No | ||
| 86 | CDC42 | 4503 8722 4504 2465 | 5208 | 0.010 | 0.5221 | No | ||
| 87 | FGF4 | 8964 | 5376 | 0.009 | 0.5132 | No | ||
| 88 | VAV2 | 5848 2670 | 5455 | 0.008 | 0.5091 | No | ||
| 89 | TMSB4X | 24005 | 5732 | 0.007 | 0.4943 | No | ||
| 90 | BDKRB2 | 21163 | 5912 | 0.006 | 0.4847 | No | ||
| 91 | MYL7 | 9437 | 5962 | 0.006 | 0.4821 | No | ||
| 92 | FGF21 | 17830 | 6170 | 0.005 | 0.4710 | No | ||
| 93 | FGF1 | 1994 23447 | 6176 | 0.005 | 0.4708 | No | ||
| 94 | ITGA11 | 6677 11449 | 6249 | 0.005 | 0.4670 | No | ||
| 95 | BAIAP2 | 449 4236 | 6433 | 0.004 | 0.4571 | No | ||
| 96 | APC | 4396 2022 | 6508 | 0.004 | 0.4532 | No | ||
| 97 | PDGFB | 22199 2311 | 6629 | 0.003 | 0.4468 | No | ||
| 98 | ITGAX | 18058 | 6630 | 0.003 | 0.4468 | No | ||
| 99 | FGF16 | 24270 | 6721 | 0.003 | 0.4420 | No | ||
| 100 | ITGA3 | 20284 | 6741 | 0.003 | 0.4410 | No | ||
| 101 | ITGB6 | 14581 2809 | 6977 | 0.002 | 0.4283 | No | ||
| 102 | CHRM2 | 10840 | 7580 | 0.001 | 0.3957 | No | ||
| 103 | PIK3CA | 9562 | 7760 | 0.001 | 0.3860 | No | ||
| 104 | FGF7 | 8965 | 7875 | 0.000 | 0.3798 | No | ||
| 105 | FGF14 | 21716 3005 3463 | 8086 | -0.000 | 0.3684 | No | ||
| 106 | FGF5 | 16785 | 8104 | -0.000 | 0.3675 | No | ||
| 107 | FGFR1 | 3789 8968 | 8129 | -0.000 | 0.3662 | No | ||
| 108 | PIP5K1C | 5250 19929 | 8142 | -0.000 | 0.3656 | No | ||
| 109 | ITGA4 | 4929 | 8201 | -0.001 | 0.3624 | No | ||
| 110 | CHRM4 | 14945 | 8268 | -0.001 | 0.3589 | No | ||
| 111 | PAK3 | 9528 | 8538 | -0.001 | 0.3443 | No | ||
| 112 | MRAS | 9409 | 8608 | -0.001 | 0.3406 | No | ||
| 113 | ITGA8 | 14685 | 8651 | -0.002 | 0.3383 | No | ||
| 114 | ARHGEF7 | 3823 | 8994 | -0.002 | 0.3198 | No | ||
| 115 | CHRM5 | 10038 | 9168 | -0.003 | 0.3105 | No | ||
| 116 | MOS | 16254 | 9303 | -0.003 | 0.3033 | No | ||
| 117 | PAK7 | 14417 | 9324 | -0.003 | 0.3023 | No | ||
| 118 | ITGA7 | 19835 | 9405 | -0.003 | 0.2980 | No | ||
| 119 | ITGB5 | 4932 | 9771 | -0.004 | 0.2783 | No | ||
| 120 | FGF8 | 23656 | 9836 | -0.005 | 0.2749 | No | ||
| 121 | FGF12 | 1723 22621 | 9991 | -0.005 | 0.2666 | No | ||
| 122 | PPP1CB | 5282 3529 3504 | 10127 | -0.005 | 0.2594 | No | ||
| 123 | ARHGEF12 | 19156 | 10291 | -0.006 | 0.2507 | No | ||
| 124 | ACTN2 | 21545 | 10336 | -0.006 | 0.2484 | No | ||
| 125 | TIAM2 | 10759 | 10533 | -0.007 | 0.2379 | No | ||
| 126 | MYH14 | 17845 | 10559 | -0.007 | 0.2366 | No | ||
| 127 | FGF11 | 20383 | 10692 | -0.007 | 0.2296 | No | ||
| 128 | NCKAP1 | 6947 | 10745 | -0.007 | 0.2269 | No | ||
| 129 | FGF10 | 4719 8962 | 10747 | -0.007 | 0.2270 | No | ||
| 130 | FGD3 | 11399 | 10867 | -0.008 | 0.2207 | No | ||
| 131 | CHRM1 | 23943 | 11348 | -0.009 | 0.1948 | No | ||
| 132 | APC2 | 10704 | 11483 | -0.010 | 0.1877 | No | ||
| 133 | WASF1 | 13514 | 11763 | -0.011 | 0.1728 | No | ||
| 134 | PFN2 | 15337 1818 | 11816 | -0.012 | 0.1702 | No | ||
| 135 | FGF20 | 18892 | 11825 | -0.012 | 0.1699 | No | ||
| 136 | PDGFRA | 16824 | 11924 | -0.012 | 0.1649 | No | ||
| 137 | FGFR3 | 8969 3566 | 12054 | -0.013 | 0.1581 | No | ||
| 138 | FGF17 | 21757 | 12265 | -0.014 | 0.1470 | No | ||
| 139 | FGF22 | 12468 7385 | 12479 | -0.016 | 0.1357 | No | ||
| 140 | FGF9 | 8966 | 12523 | -0.016 | 0.1337 | No | ||
| 141 | EGFR | 1329 20944 | 12610 | -0.017 | 0.1293 | No | ||
| 142 | FGFR4 | 3226 | 12709 | -0.018 | 0.1243 | No | ||
| 143 | BDKRB1 | 19418 | 12963 | -0.020 | 0.1109 | No | ||
| 144 | ITGB4 | 9669 159 | 12984 | -0.021 | 0.1102 | No | ||
| 145 | FGF2 | 15608 | 13003 | -0.021 | 0.1096 | No | ||
| 146 | ARPC5 | 12580 7487 | 13159 | -0.023 | 0.1016 | No | ||
| 147 | RHOA | 8624 4409 4410 | 13271 | -0.024 | 0.0960 | No | ||
| 148 | PIP5K3 | 14234 | 13388 | -0.026 | 0.0902 | No | ||
| 149 | SSH3 | 3709 10890 | 13660 | -0.030 | 0.0760 | No | ||
| 150 | FN1 | 4091 4094 971 13929 | 13809 | -0.033 | 0.0686 | No | ||
| 151 | ACTN3 | 23967 8540 | 14013 | -0.037 | 0.0582 | No | ||
| 152 | FGF3 | 17997 | 14667 | -0.058 | 0.0239 | No | ||
| 153 | VAV3 | 1774 1848 15443 | 14719 | -0.060 | 0.0222 | No | ||
| 154 | FGF23 | 17275 | 14821 | -0.065 | 0.0179 | No | ||
| 155 | PPP1R12A | 3354 19886 | 15021 | -0.076 | 0.0084 | No | ||
| 156 | ITGA9 | 19280 | 15104 | -0.082 | 0.0054 | No | ||
| 157 | ITGB3 | 20631 | 15630 | -0.137 | -0.0206 | No | ||
| 158 | ARPC1B | 3512 8628 | 15653 | -0.140 | -0.0193 | No | ||
| 159 | MAP2K2 | 19933 | 15726 | -0.151 | -0.0205 | No | ||
| 160 | MSN | 5122 | 15878 | -0.178 | -0.0255 | No | ||
| 161 | PIK3R2 | 18850 | 15900 | -0.181 | -0.0234 | No | ||
| 162 | PAK2 | 10310 5875 | 16141 | -0.223 | -0.0325 | No | ||
| 163 | DIAPH3 | 7117 | 16169 | -0.229 | -0.0298 | No | ||
| 164 | F2R | 21386 | 16204 | -0.237 | -0.0275 | No | ||
| 165 | NRAS | 5191 | 16222 | -0.243 | -0.0241 | No | ||
| 166 | PFN4 | 11798 | 16492 | -0.308 | -0.0332 | No | ||
| 167 | HRAS | 4868 | 16569 | -0.329 | -0.0314 | No | ||
| 168 | ARPC1A | 3541 16628 3519 | 16824 | -0.405 | -0.0380 | No | ||
| 169 | BCAR1 | 18741 | 17079 | -0.500 | -0.0429 | No | ||
| 170 | PPP1CC | 9609 5283 | 17139 | -0.524 | -0.0368 | No | ||
| 171 | PFN1 | 9555 | 17219 | -0.559 | -0.0311 | No | ||
| 172 | CFL1 | 4516 | 17480 | -0.687 | -0.0330 | No | ||
| 173 | ITGB7 | 22112 | 17580 | -0.749 | -0.0250 | No | ||
| 174 | PIK3R3 | 5248 | 17847 | -0.935 | -0.0228 | No | ||
| 175 | PIK3CB | 19030 | 17997 | -1.070 | -0.0118 | No | ||
| 176 | PTK2 | 22271 | 18532 | -2.547 | 0.0046 | No |