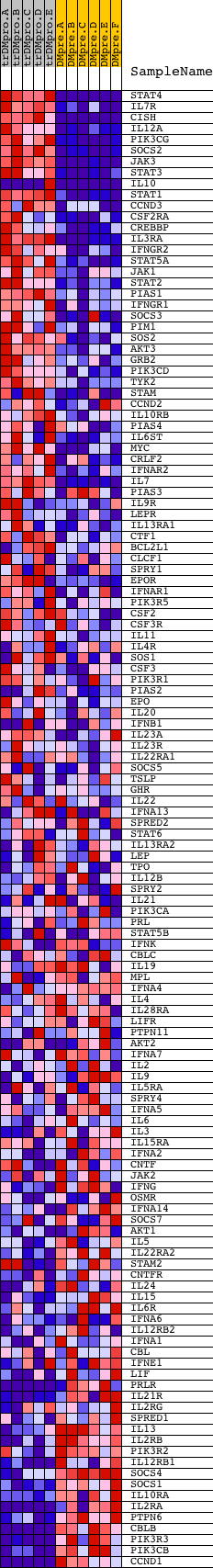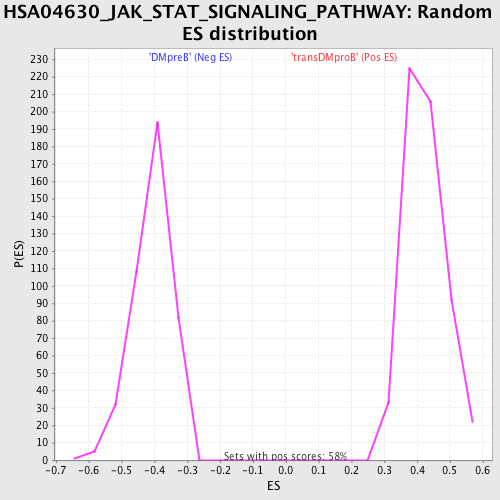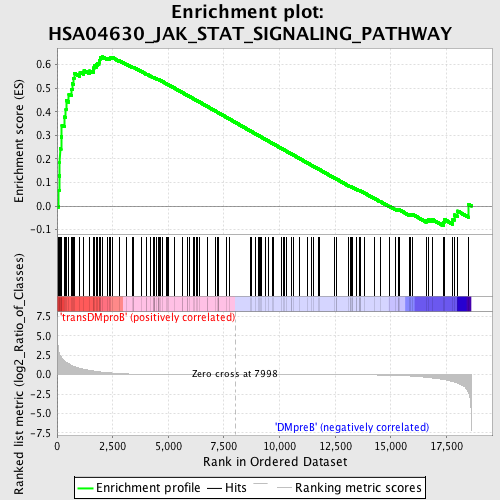
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | HSA04630_JAK_STAT_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.63300717 |
| Normalized Enrichment Score (NES) | 1.4969072 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.19843441 |
| FWER p-Value | 0.941 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | STAT4 | 14251 9907 | 80 | 2.977 | 0.0648 | Yes | ||
| 2 | IL7R | 9175 4922 | 94 | 2.773 | 0.1285 | Yes | ||
| 3 | CISH | 8743 | 122 | 2.563 | 0.1865 | Yes | ||
| 4 | IL12A | 4913 | 129 | 2.506 | 0.2444 | Yes | ||
| 5 | PIK3CG | 6635 | 176 | 2.268 | 0.2945 | Yes | ||
| 6 | SOCS2 | 5694 | 217 | 2.127 | 0.3418 | Yes | ||
| 7 | JAK3 | 9198 4936 | 320 | 1.798 | 0.3780 | Yes | ||
| 8 | STAT3 | 5525 9906 | 396 | 1.605 | 0.4112 | Yes | ||
| 9 | IL10 | 14145 1510 1553 22902 | 408 | 1.592 | 0.4476 | Yes | ||
| 10 | STAT1 | 3936 5524 | 531 | 1.404 | 0.4736 | Yes | ||
| 11 | CCND3 | 4489 4490 | 643 | 1.200 | 0.4954 | Yes | ||
| 12 | CSF2RA | 23631 | 690 | 1.122 | 0.5190 | Yes | ||
| 13 | CREBBP | 22682 8783 | 740 | 1.067 | 0.5411 | Yes | ||
| 14 | IL3RA | 22091 | 765 | 1.034 | 0.5638 | Yes | ||
| 15 | IFNGR2 | 9155 22702 | 1027 | 0.814 | 0.5686 | Yes | ||
| 16 | STAT5A | 20664 | 1207 | 0.676 | 0.5746 | Yes | ||
| 17 | JAK1 | 15827 | 1468 | 0.543 | 0.5731 | Yes | ||
| 18 | STAT2 | 19840 | 1624 | 0.469 | 0.5756 | Yes | ||
| 19 | PIAS1 | 7126 | 1633 | 0.464 | 0.5860 | Yes | ||
| 20 | IFNGR1 | 20083 | 1660 | 0.454 | 0.5951 | Yes | ||
| 21 | SOCS3 | 20131 | 1763 | 0.406 | 0.5990 | Yes | ||
| 22 | PIM1 | 23308 | 1801 | 0.391 | 0.6061 | Yes | ||
| 23 | SOS2 | 21049 | 1883 | 0.362 | 0.6101 | Yes | ||
| 24 | AKT3 | 13739 982 | 1896 | 0.357 | 0.6177 | Yes | ||
| 25 | GRB2 | 20149 | 1927 | 0.347 | 0.6242 | Yes | ||
| 26 | PIK3CD | 9563 | 1939 | 0.342 | 0.6315 | Yes | ||
| 27 | TYK2 | 12058 19215 | 2046 | 0.310 | 0.6330 | Yes | ||
| 28 | STAM | 2912 15117 | 2270 | 0.249 | 0.6267 | No | ||
| 29 | CCND2 | 16987 | 2354 | 0.232 | 0.6276 | No | ||
| 30 | IL10RB | 9169 | 2411 | 0.220 | 0.6297 | No | ||
| 31 | PIAS4 | 19681 | 2509 | 0.198 | 0.6290 | No | ||
| 32 | IL6ST | 4920 | 2808 | 0.136 | 0.6161 | No | ||
| 33 | MYC | 22465 9435 | 3120 | 0.093 | 0.6014 | No | ||
| 34 | CRLF2 | 16442 | 3390 | 0.067 | 0.5884 | No | ||
| 35 | IFNAR2 | 22705 1699 | 3428 | 0.064 | 0.5879 | No | ||
| 36 | IL7 | 4921 | 3442 | 0.063 | 0.5887 | No | ||
| 37 | PIAS3 | 15491 672 1906 | 3801 | 0.042 | 0.5703 | No | ||
| 38 | IL9R | 20505 | 4006 | 0.033 | 0.5600 | No | ||
| 39 | LEPR | 2531 2349 | 4032 | 0.032 | 0.5594 | No | ||
| 40 | IL13RA1 | 24361 | 4183 | 0.027 | 0.5519 | No | ||
| 41 | CTF1 | 18064 | 4321 | 0.024 | 0.5450 | No | ||
| 42 | BCL2L1 | 4440 2930 8652 | 4338 | 0.023 | 0.5447 | No | ||
| 43 | CLCF1 | 12160 3742 | 4367 | 0.022 | 0.5437 | No | ||
| 44 | SPRY1 | 15606 1901 | 4369 | 0.022 | 0.5441 | No | ||
| 45 | EPOR | 19204 | 4385 | 0.022 | 0.5438 | No | ||
| 46 | IFNAR1 | 22703 | 4446 | 0.020 | 0.5410 | No | ||
| 47 | PIK3R5 | 11507 | 4539 | 0.018 | 0.5365 | No | ||
| 48 | CSF2 | 20454 | 4560 | 0.018 | 0.5358 | No | ||
| 49 | CSF3R | 8804 8803 2326 4565 | 4566 | 0.018 | 0.5360 | No | ||
| 50 | IL11 | 17981 | 4592 | 0.017 | 0.5350 | No | ||
| 51 | IL4R | 18085 | 4646 | 0.016 | 0.5325 | No | ||
| 52 | SOS1 | 5476 | 4722 | 0.015 | 0.5288 | No | ||
| 53 | CSF3 | 1394 20671 | 4924 | 0.013 | 0.5182 | No | ||
| 54 | PIK3R1 | 3170 | 4940 | 0.013 | 0.5177 | No | ||
| 55 | PIAS2 | 1948 5102 | 4989 | 0.012 | 0.5154 | No | ||
| 56 | EPO | 8911 | 5254 | 0.010 | 0.5013 | No | ||
| 57 | IL20 | 13840 | 5643 | 0.007 | 0.4805 | No | ||
| 58 | IFNB1 | 15846 | 5648 | 0.007 | 0.4805 | No | ||
| 59 | IL23A | 19598 | 5855 | 0.006 | 0.4695 | No | ||
| 60 | IL23R | 17127 | 5868 | 0.006 | 0.4690 | No | ||
| 61 | IL22RA1 | 16037 | 5961 | 0.006 | 0.4641 | No | ||
| 62 | SOCS5 | 1619 23141 | 6140 | 0.005 | 0.4546 | No | ||
| 63 | TSLP | 23604 1955 | 6172 | 0.005 | 0.4531 | No | ||
| 64 | GHR | 22336 | 6281 | 0.005 | 0.4473 | No | ||
| 65 | IL22 | 11948 | 6301 | 0.005 | 0.4464 | No | ||
| 66 | IFNA13 | 15844 | 6408 | 0.004 | 0.4408 | No | ||
| 67 | SPRED2 | 8539 4332 | 6754 | 0.003 | 0.4222 | No | ||
| 68 | STAT6 | 19854 9909 | 6764 | 0.003 | 0.4218 | No | ||
| 69 | IL13RA2 | 24036 | 7116 | 0.002 | 0.4028 | No | ||
| 70 | LEP | 17505 | 7224 | 0.002 | 0.3971 | No | ||
| 71 | TPO | 21092 | 7249 | 0.002 | 0.3958 | No | ||
| 72 | IL12B | 20918 | 7262 | 0.002 | 0.3952 | No | ||
| 73 | SPRY2 | 21725 | 7275 | 0.002 | 0.3946 | No | ||
| 74 | IL21 | 12241 | 7600 | 0.001 | 0.3771 | No | ||
| 75 | PIK3CA | 9562 | 7760 | 0.001 | 0.3685 | No | ||
| 76 | PRL | 21690 | 8689 | -0.002 | 0.3183 | No | ||
| 77 | STAT5B | 20222 | 8729 | -0.002 | 0.3163 | No | ||
| 78 | IFNK | 11837 | 8932 | -0.002 | 0.3054 | No | ||
| 79 | CBLC | 17938 | 9067 | -0.003 | 0.2982 | No | ||
| 80 | IL19 | 13839 | 9097 | -0.003 | 0.2967 | No | ||
| 81 | MPL | 15780 | 9154 | -0.003 | 0.2937 | No | ||
| 82 | IFNA4 | 9150 | 9179 | -0.003 | 0.2925 | No | ||
| 83 | IL4 | 9174 | 9384 | -0.003 | 0.2815 | No | ||
| 84 | IL28RA | 10823 16038 | 9505 | -0.004 | 0.2751 | No | ||
| 85 | LIFR | 22515 9277 | 9698 | -0.004 | 0.2648 | No | ||
| 86 | PTPN11 | 5326 16391 9660 | 9713 | -0.004 | 0.2642 | No | ||
| 87 | AKT2 | 4365 4366 | 10087 | -0.005 | 0.2441 | No | ||
| 88 | IFNA7 | 9152 | 10188 | -0.006 | 0.2388 | No | ||
| 89 | IL2 | 15354 | 10226 | -0.006 | 0.2369 | No | ||
| 90 | IL9 | 21444 | 10313 | -0.006 | 0.2324 | No | ||
| 91 | IL5RA | 17051 | 10513 | -0.007 | 0.2218 | No | ||
| 92 | SPRY4 | 23448 | 10523 | -0.007 | 0.2215 | No | ||
| 93 | IFNA5 | 9151 | 10645 | -0.007 | 0.2151 | No | ||
| 94 | IL6 | 16895 | 10903 | -0.008 | 0.2014 | No | ||
| 95 | IL3 | 20453 | 11254 | -0.009 | 0.1826 | No | ||
| 96 | IL15RA | 2899 2717 15118 2792 | 11455 | -0.010 | 0.1721 | No | ||
| 97 | IFNA2 | 9149 | 11534 | -0.010 | 0.1681 | No | ||
| 98 | CNTF | 3741 8761 3680 3736 | 11752 | -0.011 | 0.1566 | No | ||
| 99 | JAK2 | 23893 9197 3706 | 11807 | -0.012 | 0.1539 | No | ||
| 100 | IFNG | 19869 | 12480 | -0.016 | 0.1180 | No | ||
| 101 | OSMR | 22333 | 12570 | -0.017 | 0.1135 | No | ||
| 102 | IFNA14 | 11917 | 12575 | -0.017 | 0.1137 | No | ||
| 103 | SOCS7 | 5308 20679 | 13105 | -0.022 | 0.0856 | No | ||
| 104 | AKT1 | 8568 | 13176 | -0.023 | 0.0823 | No | ||
| 105 | IL5 | 20884 10220 | 13198 | -0.023 | 0.0817 | No | ||
| 106 | IL22RA2 | 20082 | 13215 | -0.023 | 0.0814 | No | ||
| 107 | STAM2 | 7095 2845 | 13266 | -0.024 | 0.0793 | No | ||
| 108 | CNTFR | 2515 15906 | 13276 | -0.024 | 0.0793 | No | ||
| 109 | IL24 | 976 13841 | 13457 | -0.027 | 0.0702 | No | ||
| 110 | IL15 | 18826 3801 | 13472 | -0.027 | 0.0701 | No | ||
| 111 | IL6R | 1862 4919 | 13569 | -0.029 | 0.0656 | No | ||
| 112 | IFNA6 | 17749 | 13601 | -0.029 | 0.0646 | No | ||
| 113 | IL12RB2 | 17128 | 13627 | -0.030 | 0.0639 | No | ||
| 114 | IFNA1 | 9147 | 13637 | -0.030 | 0.0641 | No | ||
| 115 | CBL | 19154 | 13815 | -0.033 | 0.0553 | No | ||
| 116 | IFNE1 | 15842 | 14269 | -0.043 | 0.0318 | No | ||
| 117 | LIF | 956 667 20960 | 14523 | -0.051 | 0.0193 | No | ||
| 118 | PRLR | 9620 5291 | 14948 | -0.072 | -0.0020 | No | ||
| 119 | IL21R | 1398 18084 | 15221 | -0.091 | -0.0146 | No | ||
| 120 | IL2RG | 24096 | 15335 | -0.101 | -0.0183 | No | ||
| 121 | SPRED1 | 4331 | 15362 | -0.105 | -0.0173 | No | ||
| 122 | IL13 | 20461 | 15391 | -0.109 | -0.0163 | No | ||
| 123 | IL2RB | 22219 | 15852 | -0.172 | -0.0372 | No | ||
| 124 | PIK3R2 | 18850 | 15900 | -0.181 | -0.0355 | No | ||
| 125 | IL12RB1 | 3916 18582 | 15956 | -0.190 | -0.0341 | No | ||
| 126 | SOCS4 | 7424 | 16587 | -0.334 | -0.0604 | No | ||
| 127 | SOCS1 | 4522 | 16690 | -0.364 | -0.0575 | No | ||
| 128 | IL10RA | 19137 | 16879 | -0.427 | -0.0577 | No | ||
| 129 | IL2RA | 4918 | 17375 | -0.629 | -0.0699 | No | ||
| 130 | PTPN6 | 17002 | 17395 | -0.639 | -0.0561 | No | ||
| 131 | CBLB | 5531 22734 | 17786 | -0.905 | -0.0562 | No | ||
| 132 | PIK3R3 | 5248 | 17847 | -0.935 | -0.0377 | No | ||
| 133 | PIK3CB | 19030 | 17997 | -1.070 | -0.0209 | No | ||
| 134 | CCND1 | 4487 4488 8707 17535 | 18498 | -2.342 | 0.0064 | No |