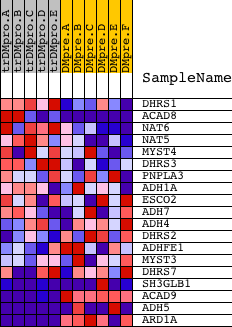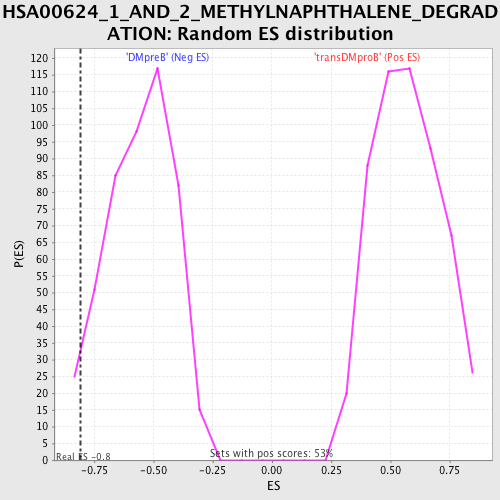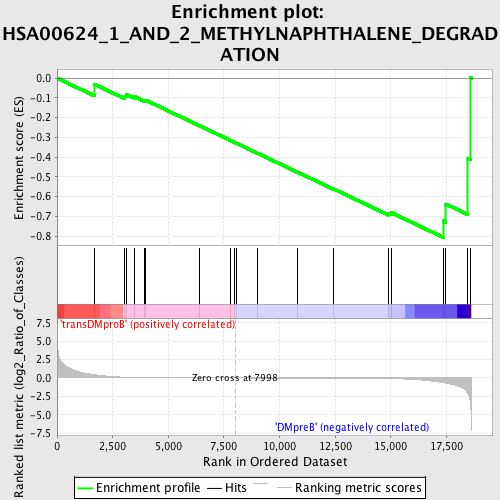
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | HSA00624_1_AND_2_METHYLNAPHTHALENE_DEGRADATION |
| Enrichment Score (ES) | -0.8070423 |
| Normalized Enrichment Score (NES) | -1.4484259 |
| Nominal p-value | 0.03805497 |
| FDR q-value | 0.33677983 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DHRS1 | 21819 | 1686 | 0.441 | -0.0318 | No | ||
| 2 | ACAD8 | 19192 3134 3032 | 3049 | 0.100 | -0.0917 | No | ||
| 3 | NAT6 | 12131 | 3114 | 0.094 | -0.0826 | No | ||
| 4 | NAT5 | 18601 7496 12597 | 3466 | 0.062 | -0.0932 | No | ||
| 5 | MYST4 | 7027 7026 12014 2739 | 3928 | 0.036 | -0.1132 | No | ||
| 6 | DHRS3 | 15997 | 3976 | 0.034 | -0.1112 | No | ||
| 7 | PNPLA3 | 22406 | 6407 | 0.004 | -0.2413 | No | ||
| 8 | ADH1A | 15406 | 7780 | 0.000 | -0.3150 | No | ||
| 9 | ESCO2 | 21781 | 7955 | 0.000 | -0.3243 | No | ||
| 10 | ADH7 | 15408 | 8062 | -0.000 | -0.3300 | No | ||
| 11 | ADH4 | 15405 | 8996 | -0.002 | -0.3799 | No | ||
| 12 | DHRS2 | 22011 | 10798 | -0.007 | -0.4757 | No | ||
| 13 | ADHFE1 | 4049 8008 | 12428 | -0.016 | -0.5612 | No | ||
| 14 | MYST3 | 18651 | 14890 | -0.069 | -0.6843 | No | ||
| 15 | DHRS7 | 2135 12338 | 15029 | -0.077 | -0.6815 | No | ||
| 16 | SH3GLB1 | 12057 7051 | 17365 | -0.626 | -0.7235 | Yes | ||
| 17 | ACAD9 | 10470 6020 | 17473 | -0.684 | -0.6379 | Yes | ||
| 18 | ADH5 | 8554 15404 | 18466 | -2.131 | -0.4067 | Yes | ||
| 19 | ARD1A | 2555 7089 12092 | 18571 | -3.105 | 0.0024 | Yes |