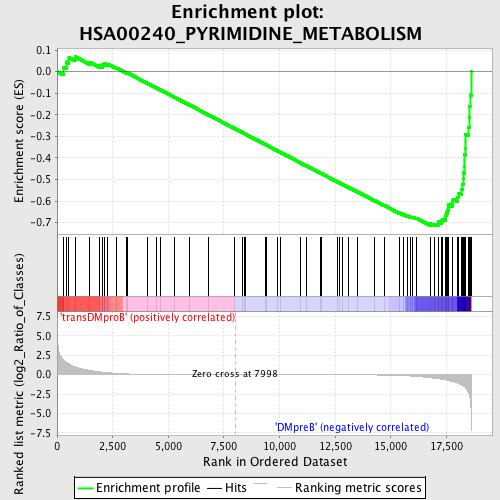
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | -0.7156337 |
| Normalized Enrichment Score (NES) | -1.6164587 |
| Nominal p-value | 0.0023148148 |
| FDR q-value | 0.05611973 |
| FWER p-Value | 0.205 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NP | 22027 9597 5273 5274 | 282 | 1.916 | 0.0213 | No | ||
| 2 | RRM2 | 5401 5400 | 406 | 1.594 | 0.0451 | No | ||
| 3 | DCK | 16808 | 533 | 1.402 | 0.0650 | No | ||
| 4 | UCK1 | 10254 5831 10253 | 810 | 0.983 | 0.0688 | No | ||
| 5 | ZNRD1 | 1491 22987 | 1460 | 0.545 | 0.0442 | No | ||
| 6 | POLD1 | 17847 | 1885 | 0.362 | 0.0282 | No | ||
| 7 | POLD4 | 12822 | 2043 | 0.311 | 0.0257 | No | ||
| 8 | POLD3 | 17742 | 2045 | 0.311 | 0.0316 | No | ||
| 9 | NT5C2 | 3768 8052 | 2123 | 0.288 | 0.0329 | No | ||
| 10 | TK2 | 18779 | 2130 | 0.286 | 0.0380 | No | ||
| 11 | RRM1 | 18163 | 2272 | 0.249 | 0.0352 | No | ||
| 12 | TK1 | 1457 10182 5762 | 2677 | 0.160 | 0.0164 | No | ||
| 13 | TXNRD1 | 19923 | 3122 | 0.093 | -0.0058 | No | ||
| 14 | POLR1A | 9749 5393 | 3151 | 0.090 | -0.0055 | No | ||
| 15 | UPP1 | 20947 1385 | 4071 | 0.030 | -0.0545 | No | ||
| 16 | CTPS2 | 12060 2571 | 4444 | 0.020 | -0.0742 | No | ||
| 17 | TYMS | 5810 5809 3606 3598 | 4667 | 0.016 | -0.0859 | No | ||
| 18 | ITPA | 9193 | 5277 | 0.010 | -0.1185 | No | ||
| 19 | POLR2C | 9750 | 5939 | 0.006 | -0.1541 | No | ||
| 20 | POLA1 | 24112 | 6822 | 0.003 | -0.2016 | No | ||
| 21 | POLE2 | 21053 | 7971 | 0.000 | -0.2635 | No | ||
| 22 | DPYS | 22305 | 8327 | -0.001 | -0.2826 | No | ||
| 23 | POLR3K | 12447 7372 | 8411 | -0.001 | -0.2871 | No | ||
| 24 | ENTPD3 | 19266 | 8468 | -0.001 | -0.2901 | No | ||
| 25 | NME7 | 926 4101 14070 3986 3946 931 | 9368 | -0.003 | -0.3385 | No | ||
| 26 | CANT1 | 13304 | 9397 | -0.003 | -0.3400 | No | ||
| 27 | ENTPD1 | 4495 | 9916 | -0.005 | -0.3678 | No | ||
| 28 | POLR2B | 16817 | 10051 | -0.005 | -0.3749 | No | ||
| 29 | AICDA | 17295 1087 | 10917 | -0.008 | -0.4214 | No | ||
| 30 | DPYD | 15437 | 11224 | -0.009 | -0.4378 | No | ||
| 31 | ENTPD8 | 12996 | 11830 | -0.012 | -0.4702 | No | ||
| 32 | UPB1 | 19988 3389 | 11898 | -0.012 | -0.4736 | No | ||
| 33 | CDA | 15702 | 12591 | -0.017 | -0.5106 | No | ||
| 34 | RRM2B | 6902 6903 11801 | 12681 | -0.018 | -0.5150 | No | ||
| 35 | NT5C1B | 21315 | 12821 | -0.019 | -0.5222 | No | ||
| 36 | UPP2 | 15009 13346 8032 | 13099 | -0.022 | -0.5367 | No | ||
| 37 | ENTPD6 | 4496 2678 | 13507 | -0.028 | -0.5581 | No | ||
| 38 | ENTPD5 | 4497 | 14287 | -0.043 | -0.5993 | No | ||
| 39 | NT5E | 19360 18702 | 14695 | -0.059 | -0.6201 | No | ||
| 40 | NUDT2 | 16239 | 15385 | -0.109 | -0.6552 | No | ||
| 41 | PNPT1 | 20936 | 15580 | -0.130 | -0.6632 | No | ||
| 42 | POLR2A | 5394 | 15732 | -0.152 | -0.6685 | No | ||
| 43 | ECGF1 | 22160 | 15899 | -0.181 | -0.6740 | No | ||
| 44 | POLR2I | 12839 | 15977 | -0.195 | -0.6744 | No | ||
| 45 | POLE4 | 12441 | 16137 | -0.223 | -0.6787 | No | ||
| 46 | POLR3A | 21900 | 16773 | -0.391 | -0.7055 | No | ||
| 47 | NT5C3 | 17136 | 16961 | -0.457 | -0.7069 | Yes | ||
| 48 | PRIM1 | 19847 | 17124 | -0.520 | -0.7057 | Yes | ||
| 49 | NME4 | 23065 1577 | 17137 | -0.524 | -0.6964 | Yes | ||
| 50 | POLE | 16755 | 17267 | -0.584 | -0.6922 | Yes | ||
| 51 | POLA2 | 23988 | 17337 | -0.614 | -0.6842 | Yes | ||
| 52 | ENTPD4 | 7449 | 17461 | -0.675 | -0.6780 | Yes | ||
| 53 | UMPS | 22606 1760 | 17470 | -0.683 | -0.6654 | Yes | ||
| 54 | POLR2J | 16672 | 17504 | -0.698 | -0.6539 | Yes | ||
| 55 | POLE3 | 7200 | 17557 | -0.740 | -0.6426 | Yes | ||
| 56 | TXNRD2 | 22829 1649 | 17575 | -0.747 | -0.6293 | Yes | ||
| 57 | POLR2G | 23753 | 17605 | -0.763 | -0.6163 | Yes | ||
| 58 | NME6 | 3139 19297 | 17766 | -0.890 | -0.6080 | Yes | ||
| 59 | NT5C | 20151 | 17793 | -0.910 | -0.5920 | Yes | ||
| 60 | NT5M | 8345 4175 | 18008 | -1.088 | -0.5828 | Yes | ||
| 61 | CAD | 16886 | 18060 | -1.150 | -0.5637 | Yes | ||
| 62 | DTYMK | 5776 | 18170 | -1.310 | -0.5446 | Yes | ||
| 63 | DHODH | 7152 | 18225 | -1.377 | -0.5212 | Yes | ||
| 64 | POLD2 | 20537 | 18251 | -1.420 | -0.4955 | Yes | ||
| 65 | POLR2K | 9413 | 18265 | -1.440 | -0.4688 | Yes | ||
| 66 | NME1 | 9467 | 18295 | -1.504 | -0.4417 | Yes | ||
| 67 | POLR2H | 10888 | 18300 | -1.513 | -0.4130 | Yes | ||
| 68 | POLR3H | 13460 8135 | 18301 | -1.515 | -0.3842 | Yes | ||
| 69 | POLR3B | 12875 | 18347 | -1.658 | -0.3550 | Yes | ||
| 70 | NME2 | 9468 | 18352 | -1.671 | -0.3234 | Yes | ||
| 71 | RFC5 | 13005 7791 | 18353 | -1.673 | -0.2915 | Yes | ||
| 72 | DCTD | 18620 | 18471 | -2.150 | -0.2568 | Yes | ||
| 73 | POLR1B | 14857 | 18527 | -2.516 | -0.2118 | Yes | ||
| 74 | CTPS | 2514 15772 | 18542 | -2.625 | -0.1625 | Yes | ||
| 75 | POLR2E | 3325 19699 | 18562 | -2.914 | -0.1080 | Yes | ||
| 76 | POLR1D | 3593 3658 16623 | 18611 | -5.818 | 0.0003 | Yes |

