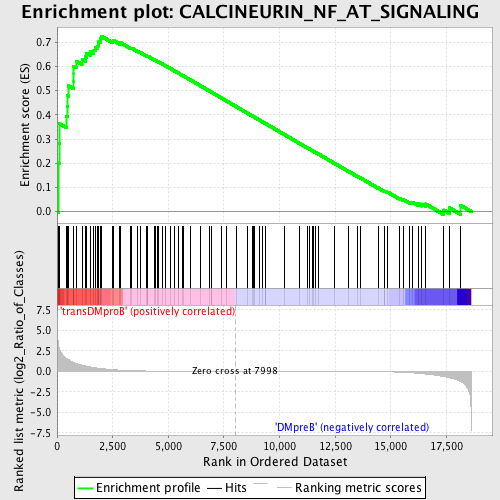
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | CALCINEURIN_NF_AT_SIGNALING |
| Enrichment Score (ES) | 0.7254018 |
| Normalized Enrichment Score (NES) | 1.6066152 |
| Nominal p-value | 0.0018315018 |
| FDR q-value | 0.06929052 |
| FWER p-Value | 0.245 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CD69 | 16979 8717 | 53 | 3.464 | 0.0994 | Yes | ||
| 2 | CDKN1A | 4511 8729 | 54 | 3.444 | 0.2012 | Yes | ||
| 3 | MAPK9 | 1233 20903 1383 | 89 | 2.817 | 0.2825 | Yes | ||
| 4 | NFKBIE | 23225 1556 | 96 | 2.766 | 0.3639 | Yes | ||
| 5 | IL10 | 14145 1510 1553 22902 | 408 | 1.592 | 0.3941 | Yes | ||
| 6 | CAMK2B | 20536 | 457 | 1.521 | 0.4364 | Yes | ||
| 7 | TGFB1 | 18332 | 479 | 1.483 | 0.4791 | Yes | ||
| 8 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 511 | 1.434 | 0.5198 | Yes | ||
| 9 | CREBBP | 22682 8783 | 740 | 1.067 | 0.5390 | Yes | ||
| 10 | NFKB2 | 23810 | 744 | 1.056 | 0.5700 | Yes | ||
| 11 | VAV1 | 23173 | 746 | 1.053 | 0.6011 | Yes | ||
| 12 | MAPK14 | 23313 | 872 | 0.931 | 0.6218 | Yes | ||
| 13 | FCGR3A | 8960 | 1120 | 0.740 | 0.6303 | Yes | ||
| 14 | NFATC3 | 5169 | 1280 | 0.631 | 0.6404 | Yes | ||
| 15 | MEF2A | 1201 5089 3492 | 1332 | 0.608 | 0.6556 | Yes | ||
| 16 | CALM3 | 8682 | 1505 | 0.524 | 0.6618 | Yes | ||
| 17 | NCK2 | 9448 | 1655 | 0.455 | 0.6672 | Yes | ||
| 18 | PPP3CC | 21763 | 1709 | 0.428 | 0.6770 | Yes | ||
| 19 | PPP3CB | 5285 | 1794 | 0.394 | 0.6841 | Yes | ||
| 20 | CALM2 | 8681 | 1840 | 0.374 | 0.6927 | Yes | ||
| 21 | GSK3B | 22761 | 1874 | 0.364 | 0.7017 | Yes | ||
| 22 | IL1B | 14431 | 1961 | 0.336 | 0.7070 | Yes | ||
| 23 | BAD | 24000 | 1967 | 0.335 | 0.7166 | Yes | ||
| 24 | RELA | 23783 | 1985 | 0.329 | 0.7254 | Yes | ||
| 25 | TRAF2 | 14657 | 2502 | 0.200 | 0.7035 | No | ||
| 26 | FCER1A | 8957 | 2519 | 0.197 | 0.7084 | No | ||
| 27 | FOS | 21202 | 2816 | 0.135 | 0.6964 | No | ||
| 28 | PAK1 | 9527 | 2858 | 0.127 | 0.6979 | No | ||
| 29 | P2RX7 | 3487 9520 487 | 3312 | 0.074 | 0.6757 | No | ||
| 30 | SP1 | 9852 | 3358 | 0.070 | 0.6753 | No | ||
| 31 | CNR1 | 4537 2362 | 3618 | 0.051 | 0.6628 | No | ||
| 32 | SFN | 7060 12062 | 3744 | 0.044 | 0.6574 | No | ||
| 33 | NFKBIB | 17906 | 4038 | 0.031 | 0.6425 | No | ||
| 34 | NFAT5 | 3921 7037 12036 | 4081 | 0.030 | 0.6411 | No | ||
| 35 | CTLA4 | 14238 | 4082 | 0.030 | 0.6420 | No | ||
| 36 | PPIA | 1284 11188 | 4398 | 0.021 | 0.6257 | No | ||
| 37 | TNF | 23004 | 4411 | 0.021 | 0.6256 | No | ||
| 38 | CEBPB | 8733 | 4530 | 0.018 | 0.6198 | No | ||
| 39 | CSF2 | 20454 | 4560 | 0.018 | 0.6188 | No | ||
| 40 | CAMK4 | 4473 | 4721 | 0.015 | 0.6106 | No | ||
| 41 | CD3G | 19139 | 4866 | 0.013 | 0.6032 | No | ||
| 42 | FKBP1B | 21116 | 5085 | 0.011 | 0.5918 | No | ||
| 43 | PPP3R1 | 9611 489 | 5297 | 0.010 | 0.5806 | No | ||
| 44 | VAV2 | 5848 2670 | 5455 | 0.008 | 0.5724 | No | ||
| 45 | IFNB1 | 15846 | 5648 | 0.007 | 0.5623 | No | ||
| 46 | MEF2B | 9377 | 5663 | 0.007 | 0.5617 | No | ||
| 47 | MYF5 | 19632 | 6004 | 0.006 | 0.5436 | No | ||
| 48 | NPPB | 15994 | 6431 | 0.004 | 0.5207 | No | ||
| 49 | PIN1 | 19548 | 6841 | 0.003 | 0.4987 | No | ||
| 50 | SP3 | 5483 | 6954 | 0.003 | 0.4927 | No | ||
| 51 | PTPRC | 5327 9662 | 6956 | 0.003 | 0.4927 | No | ||
| 52 | ICOS | 12013 | 7390 | 0.001 | 0.4694 | No | ||
| 53 | FOSL1 | 23779 | 7606 | 0.001 | 0.4578 | No | ||
| 54 | IL8RA | 13925 | 8051 | -0.000 | 0.4339 | No | ||
| 55 | ITK | 4934 | 8570 | -0.001 | 0.4060 | No | ||
| 56 | EGR2 | 8886 | 8783 | -0.002 | 0.3946 | No | ||
| 57 | CD3E | 8714 | 8819 | -0.002 | 0.3928 | No | ||
| 58 | CABIN1 | 19733 | 8892 | -0.002 | 0.3889 | No | ||
| 59 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.003 | 0.3770 | No | ||
| 60 | SLA | 5449 | 9234 | -0.003 | 0.3707 | No | ||
| 61 | IL4 | 9174 | 9384 | -0.003 | 0.3628 | No | ||
| 62 | NFATC2 | 5168 2866 | 9388 | -0.003 | 0.3627 | No | ||
| 63 | IL2 | 15354 | 10226 | -0.006 | 0.3177 | No | ||
| 64 | IL6 | 16895 | 10903 | -0.008 | 0.2815 | No | ||
| 65 | IL3 | 20453 | 11254 | -0.009 | 0.2629 | No | ||
| 66 | NFATC4 | 22002 | 11335 | -0.009 | 0.2588 | No | ||
| 67 | GATA4 | 21792 4755 | 11461 | -0.010 | 0.2524 | No | ||
| 68 | MAP2K7 | 6453 | 11501 | -0.010 | 0.2506 | No | ||
| 69 | CALM1 | 21184 | 11598 | -0.011 | 0.2457 | No | ||
| 70 | GATA3 | 9004 | 11750 | -0.011 | 0.2379 | No | ||
| 71 | IFNG | 19869 | 12480 | -0.016 | 0.1990 | No | ||
| 72 | OPRD1 | 15737 | 13098 | -0.022 | 0.1663 | No | ||
| 73 | CSNK2B | 23008 | 13521 | -0.028 | 0.1444 | No | ||
| 74 | IFNA1 | 9147 | 13637 | -0.030 | 0.1391 | No | ||
| 75 | GSK3A | 405 | 14428 | -0.048 | 0.0978 | No | ||
| 76 | VAV3 | 1774 1848 15443 | 14719 | -0.060 | 0.0840 | No | ||
| 77 | TRPV6 | 17171 | 14734 | -0.061 | 0.0850 | No | ||
| 78 | MEF2D | 9379 | 14867 | -0.067 | 0.0799 | No | ||
| 79 | IL13 | 20461 | 15391 | -0.109 | 0.0549 | No | ||
| 80 | EGR3 | 4656 | 15573 | -0.129 | 0.0489 | No | ||
| 81 | CSNK2A1 | 14797 | 15847 | -0.171 | 0.0392 | No | ||
| 82 | RPL13A | 10226 79 | 15964 | -0.192 | 0.0387 | No | ||
| 83 | MAPK8 | 6459 | 16226 | -0.245 | 0.0318 | No | ||
| 84 | JUNB | 9201 | 16390 | -0.282 | 0.0313 | No | ||
| 85 | HRAS | 4868 | 16569 | -0.329 | 0.0314 | No | ||
| 86 | IL2RA | 4918 | 17375 | -0.629 | 0.0066 | No | ||
| 87 | XPO5 | 7802 | 17624 | -0.776 | 0.0161 | No | ||
| 88 | ACTB | 8534 337 337 338 | 18146 | -1.266 | 0.0254 | No |

