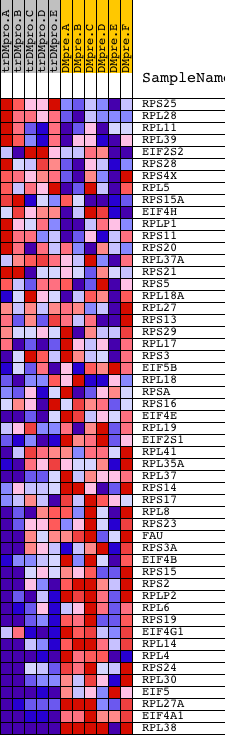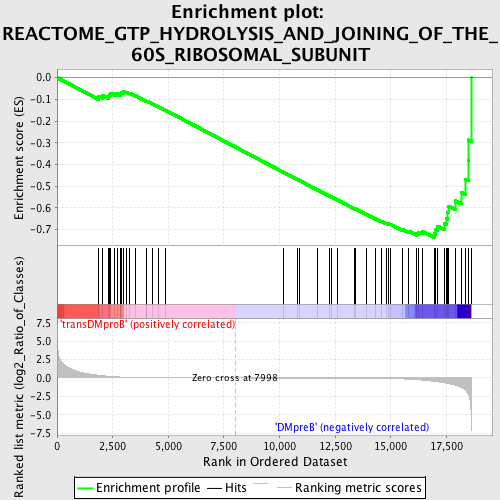
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_GTP_HYDROLYSIS_AND_JOINING_OF_THE_60S_RIBOSOMAL_SUBUNIT |
| Enrichment Score (ES) | -0.73584235 |
| Normalized Enrichment Score (NES) | -1.578877 |
| Nominal p-value | 0.004219409 |
| FDR q-value | 0.070996486 |
| FWER p-Value | 0.625 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPS25 | 13270 | 1877 | 0.364 | -0.0859 | No | ||
| 2 | RPL28 | 5392 | 2053 | 0.309 | -0.0823 | No | ||
| 3 | RPL11 | 12450 | 2322 | 0.239 | -0.0867 | No | ||
| 4 | RPL39 | 12496 | 2355 | 0.232 | -0.0787 | No | ||
| 5 | EIF2S2 | 7406 14383 | 2390 | 0.224 | -0.0711 | No | ||
| 6 | RPS28 | 12009 | 2588 | 0.181 | -0.0741 | No | ||
| 7 | RPS4X | 9756 | 2704 | 0.154 | -0.0739 | No | ||
| 8 | RPL5 | 9746 | 2846 | 0.129 | -0.0760 | No | ||
| 9 | RPS15A | 6476 | 2869 | 0.125 | -0.0720 | No | ||
| 10 | EIF4H | 16352 | 2905 | 0.118 | -0.0689 | No | ||
| 11 | RPLP1 | 3010 | 2978 | 0.108 | -0.0683 | No | ||
| 12 | RPS11 | 11317 | 2987 | 0.107 | -0.0642 | No | ||
| 13 | RPS20 | 7438 | 3110 | 0.094 | -0.0668 | No | ||
| 14 | RPL37A | 9744 | 3259 | 0.079 | -0.0714 | No | ||
| 15 | RPS21 | 12356 | 3518 | 0.058 | -0.0829 | No | ||
| 16 | RPS5 | 18391 | 4016 | 0.032 | -0.1083 | No | ||
| 17 | RPL18A | 13358 | 4031 | 0.032 | -0.1078 | No | ||
| 18 | RPL27 | 9740 | 4294 | 0.024 | -0.1208 | No | ||
| 19 | RPS13 | 12633 | 4564 | 0.018 | -0.1346 | No | ||
| 20 | RPS29 | 9754 | 4881 | 0.013 | -0.1511 | No | ||
| 21 | RPL17 | 11429 6653 | 10171 | -0.005 | -0.4358 | No | ||
| 22 | RPS3 | 6549 11295 | 10822 | -0.007 | -0.4705 | No | ||
| 23 | EIF5B | 10391 5963 | 10900 | -0.008 | -0.4743 | No | ||
| 24 | RPL18 | 450 5390 | 11686 | -0.011 | -0.5161 | No | ||
| 25 | RPSA | 19270 4984 | 12226 | -0.014 | -0.5445 | No | ||
| 26 | RPS16 | 9752 | 12339 | -0.015 | -0.5499 | No | ||
| 27 | EIF4E | 15403 1827 8890 | 12612 | -0.017 | -0.5639 | No | ||
| 28 | RPL19 | 9736 | 13379 | -0.026 | -0.6041 | No | ||
| 29 | EIF2S1 | 4658 | 13425 | -0.026 | -0.6054 | No | ||
| 30 | RPL41 | 12611 | 13922 | -0.035 | -0.6306 | No | ||
| 31 | RPL35A | 12194 | 14320 | -0.044 | -0.6502 | No | ||
| 32 | RPL37 | 12502 7421 22521 | 14583 | -0.054 | -0.6620 | No | ||
| 33 | RPS14 | 9751 | 14793 | -0.063 | -0.6706 | No | ||
| 34 | RPS17 | 9753 | 14884 | -0.068 | -0.6726 | No | ||
| 35 | RPL8 | 22437 | 14981 | -0.074 | -0.6747 | No | ||
| 36 | RPS23 | 12352 | 15534 | -0.125 | -0.6992 | No | ||
| 37 | FAU | 8954 | 15812 | -0.164 | -0.7072 | No | ||
| 38 | RPS3A | 9755 | 16163 | -0.228 | -0.7165 | No | ||
| 39 | EIF4B | 13279 7979 | 16251 | -0.250 | -0.7107 | No | ||
| 40 | RPS15 | 5396 | 16418 | -0.288 | -0.7076 | No | ||
| 41 | RPS2 | 9279 | 16943 | -0.452 | -0.7169 | Yes | ||
| 42 | RPLP2 | 7401 | 17021 | -0.478 | -0.7010 | Yes | ||
| 43 | RPL6 | 9747 | 17100 | -0.508 | -0.6838 | Yes | ||
| 44 | RPS19 | 5398 | 17418 | -0.653 | -0.6735 | Yes | ||
| 45 | EIF4G1 | 22818 | 17505 | -0.698 | -0.6489 | Yes | ||
| 46 | RPL14 | 19267 | 17549 | -0.733 | -0.6204 | Yes | ||
| 47 | RPL4 | 7499 19411 | 17597 | -0.760 | -0.5911 | Yes | ||
| 48 | RPS24 | 5399 | 17893 | -0.962 | -0.5666 | Yes | ||
| 49 | RPL30 | 9742 | 18161 | -1.285 | -0.5271 | Yes | ||
| 50 | EIF5 | 5736 | 18356 | -1.677 | -0.4672 | Yes | ||
| 51 | RPL27A | 11181 6467 18130 | 18477 | -2.186 | -0.3819 | Yes | ||
| 52 | EIF4A1 | 8889 23719 | 18492 | -2.278 | -0.2871 | Yes | ||
| 53 | RPL38 | 12562 20606 7475 | 18614 | -7.000 | 0.0001 | Yes |