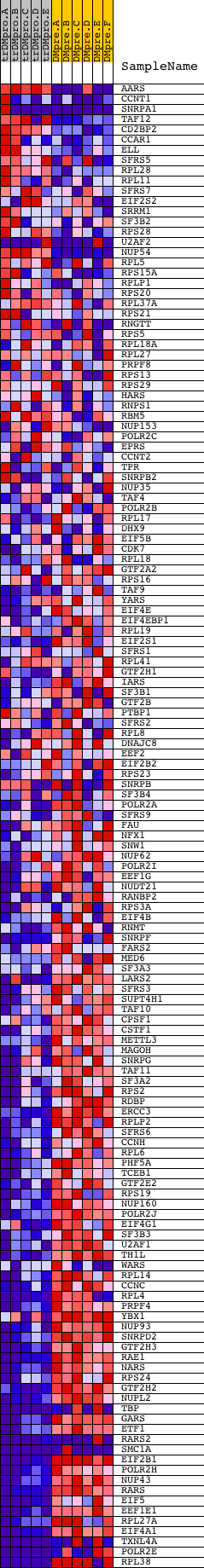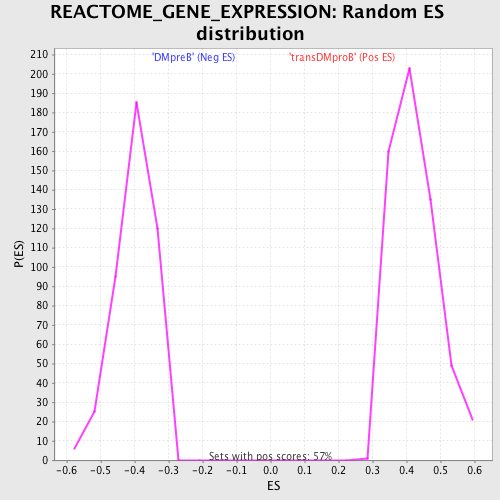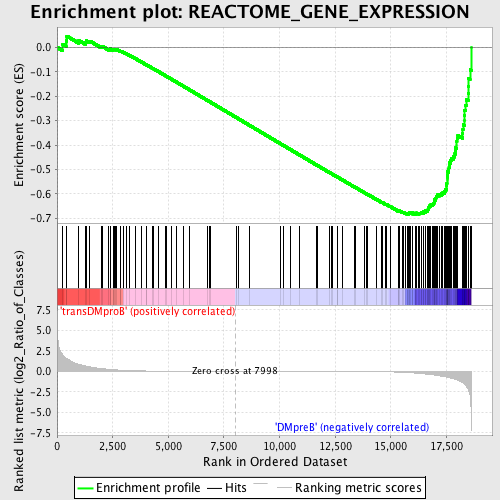
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_GENE_EXPRESSION |
| Enrichment Score (ES) | -0.6842854 |
| Normalized Enrichment Score (NES) | -1.7110479 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.01440461 |
| FWER p-Value | 0.032 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AARS | 10630 6151 | 255 | 2.001 | 0.0132 | No | ||
| 2 | CCNT1 | 22140 11607 | 405 | 1.594 | 0.0267 | No | ||
| 3 | SNRPA1 | 1373 18216 | 437 | 1.555 | 0.0461 | No | ||
| 4 | TAF12 | 2537 16058 2338 | 976 | 0.849 | 0.0284 | No | ||
| 5 | CD2BP2 | 17621 | 1288 | 0.626 | 0.0200 | No | ||
| 6 | CCAR1 | 7454 | 1318 | 0.615 | 0.0268 | No | ||
| 7 | ELL | 18586 | 1470 | 0.542 | 0.0259 | No | ||
| 8 | SFRS5 | 9808 2062 | 1972 | 0.334 | 0.0033 | No | ||
| 9 | RPL28 | 5392 | 2053 | 0.309 | 0.0032 | No | ||
| 10 | RPL11 | 12450 | 2322 | 0.239 | -0.0081 | No | ||
| 11 | SFRS7 | 22889 | 2379 | 0.226 | -0.0081 | No | ||
| 12 | EIF2S2 | 7406 14383 | 2390 | 0.224 | -0.0056 | No | ||
| 13 | SRRM1 | 6958 | 2515 | 0.198 | -0.0096 | No | ||
| 14 | SF3B2 | 23974 | 2522 | 0.196 | -0.0073 | No | ||
| 15 | RPS28 | 12009 | 2588 | 0.181 | -0.0084 | No | ||
| 16 | U2AF2 | 5813 5812 5814 | 2622 | 0.173 | -0.0078 | No | ||
| 17 | NUP54 | 11231 11232 6516 | 2686 | 0.159 | -0.0091 | No | ||
| 18 | RPL5 | 9746 | 2846 | 0.129 | -0.0160 | No | ||
| 19 | RPS15A | 6476 | 2869 | 0.125 | -0.0155 | No | ||
| 20 | RPLP1 | 3010 | 2978 | 0.108 | -0.0199 | No | ||
| 21 | RPS20 | 7438 | 3110 | 0.094 | -0.0257 | No | ||
| 22 | RPL37A | 9744 | 3259 | 0.079 | -0.0326 | No | ||
| 23 | RPS21 | 12356 | 3518 | 0.058 | -0.0458 | No | ||
| 24 | RNGTT | 10769 2354 6272 | 3773 | 0.043 | -0.0590 | No | ||
| 25 | RPS5 | 18391 | 4016 | 0.032 | -0.0716 | No | ||
| 26 | RPL18A | 13358 | 4031 | 0.032 | -0.0720 | No | ||
| 27 | RPL27 | 9740 | 4294 | 0.024 | -0.0858 | No | ||
| 28 | PRPF8 | 20780 1371 | 4344 | 0.023 | -0.0882 | No | ||
| 29 | RPS13 | 12633 | 4564 | 0.018 | -0.0998 | No | ||
| 30 | RPS29 | 9754 | 4881 | 0.013 | -0.1167 | No | ||
| 31 | HARS | 23451 | 4930 | 0.013 | -0.1191 | No | ||
| 32 | RNPS1 | 9730 23361 | 5128 | 0.011 | -0.1297 | No | ||
| 33 | RBM5 | 13507 3016 | 5365 | 0.009 | -0.1423 | No | ||
| 34 | NUP153 | 21474 | 5687 | 0.007 | -0.1596 | No | ||
| 35 | POLR2C | 9750 | 5939 | 0.006 | -0.1731 | No | ||
| 36 | EPRS | 14014 | 6779 | 0.003 | -0.2185 | No | ||
| 37 | CCNT2 | 3993 13079 | 6835 | 0.003 | -0.2214 | No | ||
| 38 | TPR | 927 4255 | 6880 | 0.003 | -0.2238 | No | ||
| 39 | SNRPB2 | 5470 | 8073 | -0.000 | -0.2883 | No | ||
| 40 | NUP35 | 12803 | 8155 | -0.000 | -0.2927 | No | ||
| 41 | TAF4 | 14319 | 8626 | -0.001 | -0.3181 | No | ||
| 42 | POLR2B | 16817 | 10051 | -0.005 | -0.3951 | No | ||
| 43 | RPL17 | 11429 6653 | 10171 | -0.005 | -0.4015 | No | ||
| 44 | DHX9 | 8845 4608 3948 | 10482 | -0.006 | -0.4182 | No | ||
| 45 | EIF5B | 10391 5963 | 10900 | -0.008 | -0.4406 | No | ||
| 46 | CDK7 | 21365 | 11644 | -0.011 | -0.4807 | No | ||
| 47 | RPL18 | 450 5390 | 11686 | -0.011 | -0.4828 | No | ||
| 48 | GTF2A2 | 10654 | 12256 | -0.014 | -0.5134 | No | ||
| 49 | RPS16 | 9752 | 12339 | -0.015 | -0.5176 | No | ||
| 50 | TAF9 | 3213 8433 | 12398 | -0.016 | -0.5206 | No | ||
| 51 | YARS | 16071 | 12601 | -0.017 | -0.5313 | No | ||
| 52 | EIF4E | 15403 1827 8890 | 12612 | -0.017 | -0.5316 | No | ||
| 53 | EIF4EBP1 | 8891 4661 | 12825 | -0.019 | -0.5428 | No | ||
| 54 | RPL19 | 9736 | 13379 | -0.026 | -0.5724 | No | ||
| 55 | EIF2S1 | 4658 | 13425 | -0.026 | -0.5745 | No | ||
| 56 | SFRS1 | 8492 | 13823 | -0.033 | -0.5955 | No | ||
| 57 | RPL41 | 12611 | 13922 | -0.035 | -0.6004 | No | ||
| 58 | GTF2H1 | 4069 18236 | 13954 | -0.036 | -0.6016 | No | ||
| 59 | IARS | 4190 8361 8362 | 14346 | -0.045 | -0.6221 | No | ||
| 60 | SF3B1 | 13954 | 14589 | -0.054 | -0.6345 | No | ||
| 61 | GTF2B | 10489 | 14615 | -0.055 | -0.6351 | No | ||
| 62 | PTBP1 | 5303 | 14763 | -0.062 | -0.6422 | No | ||
| 63 | SFRS2 | 9807 20136 | 14788 | -0.063 | -0.6426 | No | ||
| 64 | RPL8 | 22437 | 14981 | -0.074 | -0.6520 | No | ||
| 65 | DNAJC8 | 12712 | 15327 | -0.101 | -0.6694 | No | ||
| 66 | EEF2 | 8881 4654 8882 | 15348 | -0.103 | -0.6691 | No | ||
| 67 | EIF2B2 | 21204 | 15373 | -0.106 | -0.6689 | No | ||
| 68 | RPS23 | 12352 | 15534 | -0.125 | -0.6759 | No | ||
| 69 | SNRPB | 9842 5469 2736 | 15574 | -0.129 | -0.6763 | No | ||
| 70 | SF3B4 | 22269 | 15654 | -0.141 | -0.6786 | No | ||
| 71 | POLR2A | 5394 | 15732 | -0.152 | -0.6808 | No | ||
| 72 | SFRS9 | 16731 | 15758 | -0.155 | -0.6800 | No | ||
| 73 | FAU | 8954 | 15812 | -0.164 | -0.6807 | No | ||
| 74 | NFX1 | 2475 | 15830 | -0.168 | -0.6793 | No | ||
| 75 | SNW1 | 7282 | 15838 | -0.169 | -0.6774 | No | ||
| 76 | NUP62 | 9497 | 15877 | -0.178 | -0.6771 | No | ||
| 77 | POLR2I | 12839 | 15977 | -0.195 | -0.6798 | No | ||
| 78 | EEF1G | 12480 | 15985 | -0.196 | -0.6775 | No | ||
| 79 | NUDT21 | 12665 | 16111 | -0.218 | -0.6813 | Yes | ||
| 80 | RANBP2 | 20019 | 16117 | -0.220 | -0.6786 | Yes | ||
| 81 | RPS3A | 9755 | 16163 | -0.228 | -0.6780 | Yes | ||
| 82 | EIF4B | 13279 7979 | 16251 | -0.250 | -0.6793 | Yes | ||
| 83 | RNMT | 7501 | 16308 | -0.264 | -0.6788 | Yes | ||
| 84 | SNRPF | 7645 | 16356 | -0.273 | -0.6777 | Yes | ||
| 85 | FARS2 | 21666 | 16379 | -0.279 | -0.6751 | Yes | ||
| 86 | MED6 | 21028 | 16461 | -0.298 | -0.6754 | Yes | ||
| 87 | SF3A3 | 16091 | 16480 | -0.305 | -0.6723 | Yes | ||
| 88 | LARS2 | 19255 | 16561 | -0.327 | -0.6722 | Yes | ||
| 89 | SFRS3 | 5428 23312 | 16566 | -0.328 | -0.6680 | Yes | ||
| 90 | SUPT4H1 | 9938 | 16665 | -0.359 | -0.6684 | Yes | ||
| 91 | TAF10 | 10785 | 16670 | -0.360 | -0.6638 | Yes | ||
| 92 | CPSF1 | 22241 | 16700 | -0.367 | -0.6604 | Yes | ||
| 93 | CSTF1 | 12515 | 16705 | -0.367 | -0.6556 | Yes | ||
| 94 | METTL3 | 21841 | 16720 | -0.371 | -0.6514 | Yes | ||
| 95 | MAGOH | 2386 16148 | 16755 | -0.386 | -0.6480 | Yes | ||
| 96 | SNRPG | 12622 | 16801 | -0.397 | -0.6451 | Yes | ||
| 97 | TAF11 | 12733 | 16877 | -0.426 | -0.6434 | Yes | ||
| 98 | SF3A2 | 19938 | 16915 | -0.442 | -0.6394 | Yes | ||
| 99 | RPS2 | 9279 | 16943 | -0.452 | -0.6347 | Yes | ||
| 100 | RDBP | 6589 | 16947 | -0.453 | -0.6288 | Yes | ||
| 101 | ERCC3 | 23605 | 16956 | -0.457 | -0.6230 | Yes | ||
| 102 | RPLP2 | 7401 | 17021 | -0.478 | -0.6200 | Yes | ||
| 103 | SFRS6 | 14751 | 17031 | -0.480 | -0.6140 | Yes | ||
| 104 | CCNH | 7322 | 17056 | -0.491 | -0.6087 | Yes | ||
| 105 | RPL6 | 9747 | 17100 | -0.508 | -0.6042 | Yes | ||
| 106 | PHF5A | 22194 | 17185 | -0.544 | -0.6014 | Yes | ||
| 107 | TCEB1 | 13995 | 17277 | -0.590 | -0.5983 | Yes | ||
| 108 | GTF2E2 | 18635 | 17340 | -0.615 | -0.5933 | Yes | ||
| 109 | RPS19 | 5398 | 17418 | -0.653 | -0.5887 | Yes | ||
| 110 | NUP160 | 14957 | 17457 | -0.672 | -0.5817 | Yes | ||
| 111 | POLR2J | 16672 | 17504 | -0.698 | -0.5747 | Yes | ||
| 112 | EIF4G1 | 22818 | 17505 | -0.698 | -0.5653 | Yes | ||
| 113 | SF3B3 | 18746 | 17517 | -0.706 | -0.5563 | Yes | ||
| 114 | U2AF1 | 8431 | 17527 | -0.715 | -0.5472 | Yes | ||
| 115 | TH1L | 14715 | 17528 | -0.716 | -0.5375 | Yes | ||
| 116 | WARS | 2085 20984 | 17532 | -0.718 | -0.5280 | Yes | ||
| 117 | RPL14 | 19267 | 17549 | -0.733 | -0.5189 | Yes | ||
| 118 | CCNC | 16263 | 17555 | -0.736 | -0.5092 | Yes | ||
| 119 | RPL4 | 7499 19411 | 17597 | -0.760 | -0.5012 | Yes | ||
| 120 | PRPF4 | 7655 12845 | 17613 | -0.769 | -0.4916 | Yes | ||
| 121 | YBX1 | 5929 2389 | 17628 | -0.778 | -0.4818 | Yes | ||
| 122 | NUP93 | 7762 | 17648 | -0.802 | -0.4720 | Yes | ||
| 123 | SNRPD2 | 8412 | 17693 | -0.836 | -0.4631 | Yes | ||
| 124 | GTF2H3 | 5551 | 17720 | -0.851 | -0.4530 | Yes | ||
| 125 | RAE1 | 12395 | 17803 | -0.914 | -0.4451 | Yes | ||
| 126 | NARS | 23418 | 17879 | -0.953 | -0.4363 | Yes | ||
| 127 | RPS24 | 5399 | 17893 | -0.962 | -0.4240 | Yes | ||
| 128 | GTF2H2 | 6236 | 17906 | -0.975 | -0.4115 | Yes | ||
| 129 | NUPL2 | 6072 | 17965 | -1.029 | -0.4007 | Yes | ||
| 130 | TBP | 671 1554 | 17966 | -1.031 | -0.3868 | Yes | ||
| 131 | GARS | 17438 | 17975 | -1.042 | -0.3731 | Yes | ||
| 132 | ETF1 | 23467 | 18013 | -1.092 | -0.3603 | Yes | ||
| 133 | RARS2 | 16247 2490 2411 | 18226 | -1.379 | -0.3532 | Yes | ||
| 134 | SMC1A | 2572 24225 6279 10780 | 18238 | -1.400 | -0.3349 | Yes | ||
| 135 | EIF2B1 | 16368 3458 | 18250 | -1.416 | -0.3163 | Yes | ||
| 136 | POLR2H | 10888 | 18300 | -1.513 | -0.2985 | Yes | ||
| 137 | NUP43 | 20094 | 18304 | -1.526 | -0.2780 | Yes | ||
| 138 | RARS | 20496 | 18316 | -1.556 | -0.2576 | Yes | ||
| 139 | EIF5 | 5736 | 18356 | -1.677 | -0.2370 | Yes | ||
| 140 | EEF1E1 | 21488 | 18400 | -1.815 | -0.2148 | Yes | ||
| 141 | RPL27A | 11181 6467 18130 | 18477 | -2.186 | -0.1894 | Yes | ||
| 142 | EIF4A1 | 8889 23719 | 18492 | -2.278 | -0.1594 | Yes | ||
| 143 | TXNL4A | 6567 11329 | 18503 | -2.361 | -0.1280 | Yes | ||
| 144 | POLR2E | 3325 19699 | 18562 | -2.914 | -0.0918 | Yes | ||
| 145 | RPL38 | 12562 20606 7475 | 18614 | -7.000 | 0.0001 | Yes |