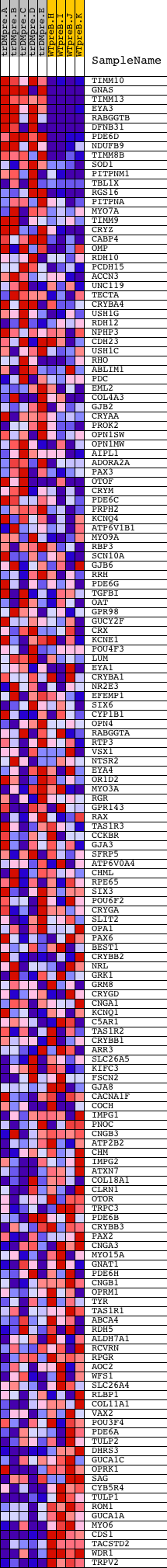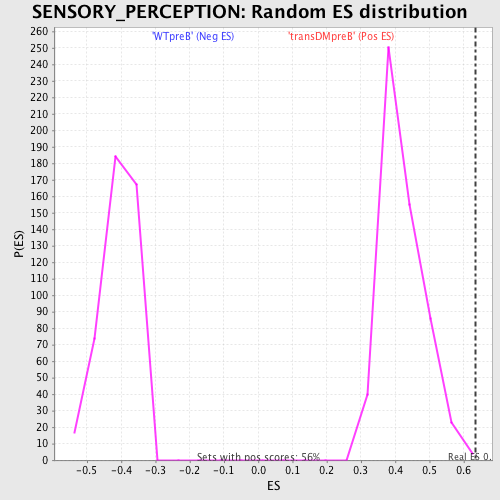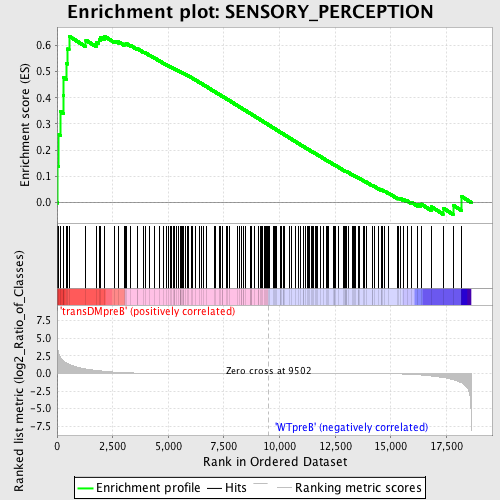
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | SENSORY_PERCEPTION |
| Enrichment Score (ES) | 0.6351293 |
| Normalized Enrichment Score (NES) | 1.5129944 |
| Nominal p-value | 0.005376344 |
| FDR q-value | 0.16760322 |
| FWER p-Value | 0.974 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM10 | 11390 2158 | 30 | 3.408 | 0.1372 | Yes | ||
| 2 | GNAS | 9025 2963 2752 | 54 | 3.075 | 0.2612 | Yes | ||
| 3 | TIMM13 | 14974 | 149 | 2.246 | 0.3476 | Yes | ||
| 4 | EYA3 | 8936 2352 | 286 | 1.699 | 0.4095 | Yes | ||
| 5 | RABGGTB | 15134 | 299 | 1.660 | 0.4764 | Yes | ||
| 6 | DFNB31 | 13139 | 410 | 1.502 | 0.5316 | Yes | ||
| 7 | PDE6D | 13897 3945 | 467 | 1.413 | 0.5861 | Yes | ||
| 8 | NDUFB9 | 12306 | 537 | 1.294 | 0.6351 | Yes | ||
| 9 | TIMM8B | 11391 | 1293 | 0.625 | 0.6197 | No | ||
| 10 | SOD1 | 9846 | 1769 | 0.431 | 0.6115 | No | ||
| 11 | PITPNM1 | 23770 | 1882 | 0.382 | 0.6210 | No | ||
| 12 | TBL1X | 5628 | 1958 | 0.358 | 0.6315 | No | ||
| 13 | RGS16 | 14094 | 2122 | 0.306 | 0.6351 | No | ||
| 14 | PITPNA | 20778 | 2600 | 0.173 | 0.6163 | No | ||
| 15 | MYO7A | 9439 | 2745 | 0.150 | 0.6146 | No | ||
| 16 | TIMM9 | 19686 | 3007 | 0.107 | 0.6048 | No | ||
| 17 | CRYZ | 1873 15387 1927 | 3074 | 0.097 | 0.6052 | No | ||
| 18 | CABP4 | 13125 | 3118 | 0.092 | 0.6066 | No | ||
| 19 | OMP | 5210 | 3300 | 0.074 | 0.5998 | No | ||
| 20 | RDH10 | 8272 | 3595 | 0.054 | 0.5861 | No | ||
| 21 | PCDH15 | 4433 | 3622 | 0.053 | 0.5868 | No | ||
| 22 | ACCN3 | 3532 3647 | 3889 | 0.042 | 0.5741 | No | ||
| 23 | UNC119 | 20758 | 3954 | 0.039 | 0.5722 | No | ||
| 24 | TECTA | 19159 | 4159 | 0.033 | 0.5625 | No | ||
| 25 | CRYBA4 | 21596 | 4394 | 0.028 | 0.5510 | No | ||
| 26 | USH1G | 20153 | 4597 | 0.024 | 0.5410 | No | ||
| 27 | RDH12 | 21227 | 4779 | 0.022 | 0.5321 | No | ||
| 28 | NPHP3 | 120 19337 | 4918 | 0.020 | 0.5254 | No | ||
| 29 | CDH23 | 3369 19759 3442 | 4999 | 0.019 | 0.5218 | No | ||
| 30 | USH1C | 1471 17821 | 5092 | 0.018 | 0.5176 | No | ||
| 31 | RHO | 17314 | 5119 | 0.017 | 0.5169 | No | ||
| 32 | ABLIM1 | 3717 3766 3679 | 5229 | 0.016 | 0.5116 | No | ||
| 33 | PDC | 4046 14102 | 5270 | 0.016 | 0.5101 | No | ||
| 34 | EML2 | 3240 | 5271 | 0.016 | 0.5108 | No | ||
| 35 | COL4A3 | 8766 | 5370 | 0.015 | 0.5061 | No | ||
| 36 | GJB2 | 9018 21806 | 5441 | 0.015 | 0.5029 | No | ||
| 37 | CRYAA | 23298 | 5542 | 0.014 | 0.4980 | No | ||
| 38 | PROK2 | 17057 | 5549 | 0.014 | 0.4982 | No | ||
| 39 | OPN1SW | 17196 | 5553 | 0.014 | 0.4986 | No | ||
| 40 | OPN1MW | 24302 | 5610 | 0.013 | 0.4961 | No | ||
| 41 | AIPL1 | 4314 | 5644 | 0.013 | 0.4949 | No | ||
| 42 | ADORA2A | 8557 | 5666 | 0.013 | 0.4943 | No | ||
| 43 | PAX3 | 13910 | 5690 | 0.013 | 0.4935 | No | ||
| 44 | OTOF | 16578 3599 | 5763 | 0.012 | 0.4901 | No | ||
| 45 | CRYM | 17655 | 5880 | 0.011 | 0.4843 | No | ||
| 46 | PDE6C | 8496 23868 | 5889 | 0.011 | 0.4843 | No | ||
| 47 | PRPH2 | 23214 | 6035 | 0.011 | 0.4769 | No | ||
| 48 | KCNQ4 | 12247 2533 | 6074 | 0.010 | 0.4753 | No | ||
| 49 | ATP6V1B1 | 8499 | 6225 | 0.010 | 0.4676 | No | ||
| 50 | MYO9A | 11325 | 6398 | 0.009 | 0.4586 | No | ||
| 51 | RBP3 | 22047 | 6489 | 0.009 | 0.4541 | No | ||
| 52 | SCN10A | 18969 | 6591 | 0.008 | 0.4489 | No | ||
| 53 | GJB6 | 21805 | 6734 | 0.008 | 0.4416 | No | ||
| 54 | RRH | 15168 | 7055 | 0.007 | 0.4245 | No | ||
| 55 | PDE6G | 20119 | 7120 | 0.006 | 0.4213 | No | ||
| 56 | TGFBI | 3181 21622 | 7317 | 0.006 | 0.4109 | No | ||
| 57 | OAT | 17594 | 7328 | 0.006 | 0.4106 | No | ||
| 58 | GPR98 | 21400 | 7426 | 0.005 | 0.4055 | No | ||
| 59 | GUCY2F | 24045 | 7454 | 0.005 | 0.4043 | No | ||
| 60 | CRX | 17961 4694 | 7608 | 0.005 | 0.3962 | No | ||
| 61 | KCNE1 | 9206 22537 4942 | 7652 | 0.005 | 0.3941 | No | ||
| 62 | POU4F3 | 9604 | 7727 | 0.004 | 0.3902 | No | ||
| 63 | LUM | 19896 | 8106 | 0.004 | 0.3699 | No | ||
| 64 | EYA1 | 4695 4061 | 8207 | 0.003 | 0.3646 | No | ||
| 65 | CRYBA1 | 20341 1409 | 8297 | 0.003 | 0.3599 | No | ||
| 66 | NR2E3 | 3084 | 8303 | 0.003 | 0.3598 | No | ||
| 67 | EFEMP1 | 20937 | 8369 | 0.003 | 0.3563 | No | ||
| 68 | SIX6 | 21247 | 8379 | 0.003 | 0.3560 | No | ||
| 69 | CYP1B1 | 4581 | 8475 | 0.003 | 0.3509 | No | ||
| 70 | OPN4 | 21876 | 8677 | 0.002 | 0.3401 | No | ||
| 71 | RABGGTA | 2669 21820 | 8706 | 0.002 | 0.3387 | No | ||
| 72 | RTP3 | 18983 | 8746 | 0.002 | 0.3366 | No | ||
| 73 | VSX1 | 2766 14398 | 8754 | 0.002 | 0.3363 | No | ||
| 74 | NTSR2 | 21310 | 8864 | 0.002 | 0.3305 | No | ||
| 75 | EYA4 | 8937 | 9047 | 0.001 | 0.3207 | No | ||
| 76 | OR1D2 | 6420 | 9049 | 0.001 | 0.3207 | No | ||
| 77 | MYO3A | 10065 | 9138 | 0.001 | 0.3159 | No | ||
| 78 | RGR | 21874 3042 | 9194 | 0.001 | 0.3130 | No | ||
| 79 | GPR143 | 24224 | 9218 | 0.001 | 0.3118 | No | ||
| 80 | RAX | 5361 | 9315 | 0.000 | 0.3066 | No | ||
| 81 | TAS1R3 | 15643 | 9355 | 0.000 | 0.3045 | No | ||
| 82 | CCKBR | 8706 | 9431 | 0.000 | 0.3004 | No | ||
| 83 | GJA3 | 21807 | 9474 | 0.000 | 0.2982 | No | ||
| 84 | SFRP5 | 23674 | 9507 | -0.000 | 0.2964 | No | ||
| 85 | ATP6V0A4 | 8934 | 9568 | -0.000 | 0.2932 | No | ||
| 86 | CHML | 13741 | 9710 | -0.001 | 0.2856 | No | ||
| 87 | RPE65 | 15382 | 9755 | -0.001 | 0.2832 | No | ||
| 88 | SIX3 | 5444 | 9828 | -0.001 | 0.2794 | No | ||
| 89 | POU6F2 | 21539 | 9871 | -0.001 | 0.2771 | No | ||
| 90 | CRYGA | 8794 | 10030 | -0.001 | 0.2686 | No | ||
| 91 | SLIT2 | 5456 | 10042 | -0.001 | 0.2681 | No | ||
| 92 | OPA1 | 13169 22800 7894 | 10063 | -0.001 | 0.2671 | No | ||
| 93 | PAX6 | 5223 | 10181 | -0.002 | 0.2608 | No | ||
| 94 | BEST1 | 23747 | 10197 | -0.002 | 0.2600 | No | ||
| 95 | CRYBB2 | 16433 | 10201 | -0.002 | 0.2600 | No | ||
| 96 | NRL | 9484 | 10445 | -0.002 | 0.2469 | No | ||
| 97 | GRK1 | 18677 | 10524 | -0.003 | 0.2428 | No | ||
| 98 | GRM8 | 4805 | 10700 | -0.003 | 0.2334 | No | ||
| 99 | CRYGD | 8797 | 10827 | -0.003 | 0.2267 | No | ||
| 100 | CNGA1 | 8757 | 10954 | -0.004 | 0.2200 | No | ||
| 101 | KCNQ1 | 18001 4948 | 11067 | -0.004 | 0.2141 | No | ||
| 102 | C5AR1 | 17957 | 11086 | -0.004 | 0.2133 | No | ||
| 103 | TAS1R2 | 16010 | 11157 | -0.004 | 0.2097 | No | ||
| 104 | CRYBB1 | 16748 | 11268 | -0.005 | 0.2039 | No | ||
| 105 | ARR3 | 24282 9305 | 11319 | -0.005 | 0.2014 | No | ||
| 106 | SLC26A5 | 3667 16593 | 11356 | -0.005 | 0.1997 | No | ||
| 107 | KIFC3 | 9227 | 11432 | -0.005 | 0.1958 | No | ||
| 108 | FSCN2 | 20569 | 11496 | -0.005 | 0.1926 | No | ||
| 109 | GJA8 | 15234 | 11543 | -0.005 | 0.1903 | No | ||
| 110 | CACNA1F | 12054 24393 | 11613 | -0.006 | 0.1868 | No | ||
| 111 | COCH | 21277 | 11660 | -0.006 | 0.1846 | No | ||
| 112 | IMPG1 | 19052 | 11692 | -0.006 | 0.1831 | No | ||
| 113 | PNOC | 21783 | 11833 | -0.006 | 0.1758 | No | ||
| 114 | CNGB3 | 16267 | 11969 | -0.007 | 0.1688 | No | ||
| 115 | ATP2B2 | 17038 | 12109 | -0.007 | 0.1615 | No | ||
| 116 | CHM | 4521 | 12142 | -0.008 | 0.1601 | No | ||
| 117 | IMPG2 | 22731 | 12183 | -0.008 | 0.1583 | No | ||
| 118 | ATXN7 | 3105 | 12444 | -0.009 | 0.1445 | No | ||
| 119 | COL18A1 | 19719 3360 | 12446 | -0.009 | 0.1449 | No | ||
| 120 | CLRN1 | 1828 6025 10474 | 12469 | -0.009 | 0.1440 | No | ||
| 121 | OTOR | 14825 | 12533 | -0.009 | 0.1410 | No | ||
| 122 | TRPC3 | 15355 | 12642 | -0.010 | 0.1356 | No | ||
| 123 | PDE6B | 16760 | 12880 | -0.011 | 0.1232 | No | ||
| 124 | CRYBB3 | 16432 | 12937 | -0.011 | 0.1206 | No | ||
| 125 | PAX2 | 9530 24416 | 12956 | -0.012 | 0.1201 | No | ||
| 126 | CNGA3 | 8758 4535 | 12963 | -0.012 | 0.1202 | No | ||
| 127 | MYO15A | 1242 20860 | 13006 | -0.012 | 0.1184 | No | ||
| 128 | GNAT1 | 3027 19000 | 13013 | -0.012 | 0.1186 | No | ||
| 129 | PDE6H | 17257 | 13103 | -0.013 | 0.1143 | No | ||
| 130 | CNGB1 | 18784 | 13263 | -0.014 | 0.1062 | No | ||
| 131 | OPRM1 | 5212 | 13308 | -0.014 | 0.1044 | No | ||
| 132 | TYR | 17763 | 13346 | -0.014 | 0.1030 | No | ||
| 133 | TAS1R1 | 15657 | 13410 | -0.015 | 0.1002 | No | ||
| 134 | ABCA4 | 8508 15434 | 13526 | -0.016 | 0.0946 | No | ||
| 135 | RDH5 | 19599 8409 4214 4215 19587 | 13535 | -0.016 | 0.0948 | No | ||
| 136 | ALDH7A1 | 23435 | 13579 | -0.016 | 0.0932 | No | ||
| 137 | RCVRN | 20835 | 13775 | -0.019 | 0.0834 | No | ||
| 138 | RPGR | 9734 | 13833 | -0.019 | 0.0811 | No | ||
| 139 | AOC2 | 20658 1256 | 13888 | -0.020 | 0.0790 | No | ||
| 140 | WFS1 | 16548 | 14173 | -0.024 | 0.0646 | No | ||
| 141 | SLC26A4 | 21089 | 14190 | -0.025 | 0.0647 | No | ||
| 142 | RLBP1 | 17790 | 14255 | -0.026 | 0.0623 | No | ||
| 143 | COL11A1 | 15442 1937 4540 8762 | 14466 | -0.030 | 0.0521 | No | ||
| 144 | VAX2 | 17390 | 14561 | -0.032 | 0.0484 | No | ||
| 145 | POU3F4 | 347 | 14572 | -0.033 | 0.0491 | No | ||
| 146 | PDE6A | 10345 | 14615 | -0.034 | 0.0482 | No | ||
| 147 | TULP2 | 18247 | 14722 | -0.037 | 0.0440 | No | ||
| 148 | DHRS3 | 15997 | 14906 | -0.043 | 0.0358 | No | ||
| 149 | GUCA1C | 4761 | 15313 | -0.062 | 0.0164 | No | ||
| 150 | OPRK1 | 14300 | 15356 | -0.066 | 0.0168 | No | ||
| 151 | SAG | 5404 | 15434 | -0.072 | 0.0155 | No | ||
| 152 | CYB5R4 | 6475 19361 | 15556 | -0.082 | 0.0123 | No | ||
| 153 | TULP1 | 1555 23052 | 15730 | -0.098 | 0.0070 | No | ||
| 154 | ROM1 | 23751 | 15914 | -0.123 | 0.0021 | No | ||
| 155 | GUCA1A | 22952 | 16217 | -0.189 | -0.0066 | No | ||
| 156 | MYO6 | 19368 | 16356 | -0.219 | -0.0052 | No | ||
| 157 | CDS1 | 16779 | 16827 | -0.363 | -0.0158 | No | ||
| 158 | TACSTD2 | 17126 | 17349 | -0.572 | -0.0208 | No | ||
| 159 | WDR1 | 16544 3592 | 17801 | -0.867 | -0.0099 | No | ||
| 160 | TRPV2 | 20849 | 18179 | -1.325 | 0.0237 | No |